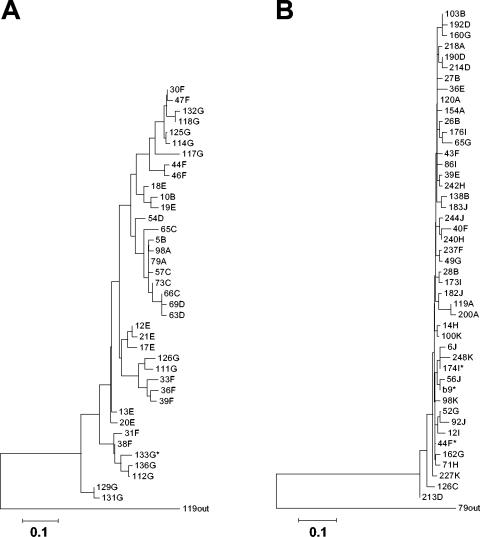
FIG. 2.
Phylogenetic analysis of HCV HVR1 sequences derived from peripheral blood or liver samples obtained from coinfected children. (A) Case 1. A total of 115 independently derived HVR1 sequences (mean of 16.4 clones per time point; n = 13 to 20) were analyzed using the neighbor-joining method as described in the text. A transition/transversion ratio of 0.5 was used, and 500 bootstrap resamplings were performed. Letters correspond to the specific time point after birth at which sequences were isolated (A, 0.13 years; B, 0.14 years; C, 1.25 years; D, 1.56 years; E, 3.02 years; F, 4.88 years; G, 5.87 years). The asterisk indicates a sequence identical to that of a liver-derived variant. (B) Case 2. A total of 212 independently derived HVR1 sequences (mean of 21.2 clones per time point; n = 16 to 24) were analyzed as described above. Letters correspond to the specific time points after birth at which sequences were isolated (A, 0.21 years; B, 2.07 years; C, 2.30 years; D, 3.60 years; E, 3.83 years; F, 4.29 years; G, 7.07 years; H, 7.46 years; I, 7.76 years; J, 10.03 years; K, 12.65 years). Asterisks indicate the sequence of a liver-derived variant (b9) or sequences identical thereto. Predominant variants from the first time points were used as reciprocal outgroups in both analyses. The scale bars represent 0.1 nucleotide substitutions per site.