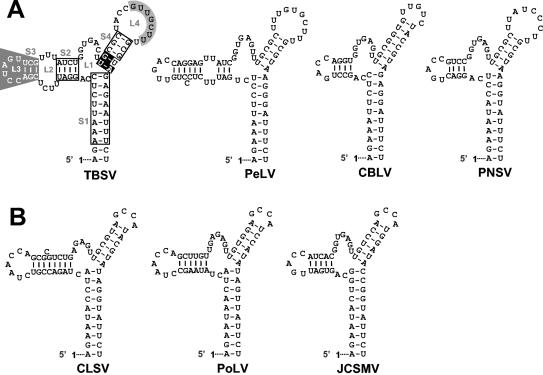
FIG. 9.
RNA secondary structure comparison of the TBSV TSD with those from recently sequenced tombusviruses (A) and aureusviruses (B). In panel A, the nucleotides in open boxes indicate positions where compensatory mutational analyses from this and previous (32) studies have indicated that secondary structure is important. S4 nucleotides, in white on black, are entirely conserved within the genera Tombusvirus and Aureusvirus, and their identities are important for TSD activity. L4 nucleotides that interact with the adjacent DSD to form PK-TD1 are in black on grey, while SL3 nucleotides involved in a long-distance RNA-RNA interaction with the 3′-proximal translational enhancer are in white on grey (6, 23). For the aureusvirus TSDs shown in panel B, corresponding local pseudoknot and long-distance interactions have been predicted and presented elsewhere (6, 23). The virus abbreviations are as follows: PeLV, Pear latent virus; CBLV, Cucumber Bulgarian latent virus; PNSV, Pelargonium necrotic spot virus; CLSV, Cucumber leaf spot virus; PoLV, Pothos latent virus; and JCSMV, Johngrass chlorotic stripe mosaic virus.