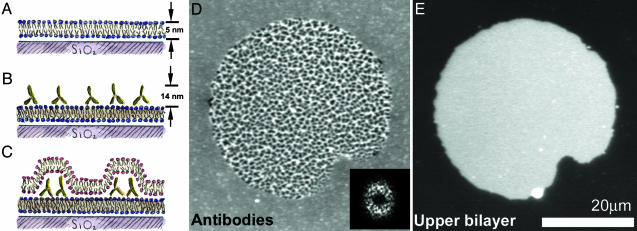
Fig. 1.
Protein patterns at lipid bilayer junctions. (A–C) Schematic (side view) of the experimental setup. (A) A supported lipid bilayer, with ≈1% biotinylated lipid, self-assembles on a glass substrate. (B) Antibiotin antibodies bind to the supported bilayer, forming a layer of fluid, membrane-bound protein. (C) Giant lipid vesicles tens of micrometers in diameter (schematic not shown) are introduced to the supported bilayer/antibody system. The giant vesicles rupture, creating a bilayer-bilayer junction. Interbilayer adhesion leads to reorganization of the proteins into dense and sparse regions. (D) Fluorescence image (top view) of FITC-labeled antibiotin; the central region, corresponding to the bilayer-bilayer junction, is patterned into zones of high and low density. The protein patterns often exhibit considerable regularity; the 2D Fourier transform (2DFT) shows a broad ring corresponding to a spatial periodicity ≈1–2 μm in wavelength. (Inset) The squared magnitude of the 2DFT. (E) Fluorescence image (top view) of the Texas red-labeled upper bilayer; its finite extent defines the area of the intermembrane junction. The system is in an aqueous solution of 3 mM NaCl.