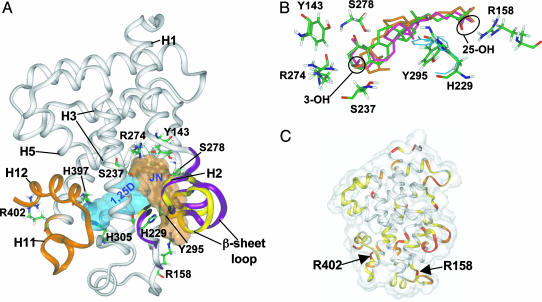
Fig. 2.
VDR molecular models depicting the G and A pockets. (A) Ribbon diagrams of the assemblies resulting from molecular modeling of the VDR LBD (x-ray coordinates; Protein Data Bank ID code 1DB1) with 6-s-trans 1,25D in the G pocket (cyan Connolly surface) and JN docked in the A pocket (light-brown Connolly surface). The orange ribbon, H11 and H12, is shown with H12 in the closed, transcriptionally active conformation. The yellow ribbons indicate the orientation of the H2/β-sheet region in VDRwt, whereas the purple ribbons indicate their final position after JN was docked in the A pocket. Important amino acid residues discussed in the text are labeled. (B) Superimposition of the VDR amide backbone atoms of the 1,25D α/β-chair and JN A pocket models. JN's oxygen atoms are red and carbon atoms are green (Colat colors). The α-chair form of 1,25D is orange and the β-chair is pink. The starting orientations of Y295 and H229 are indicated by the thin, cyan wireframe and their final positions are indicated by the thicker, Colat-colored wireframe. (C) Superimposition of the 1,25D/VDR G pocket and the 6-s-cis 1,25D/VDR A pocket (backbone rms = 1.18 Å) models rendered to show the VDR LBD Connolly surface (transparent). The ribbon diagram is color-coded to represent the degree of movement observed when the two models are compared. The red regions indicate >3.0 Å, orange between 2.0 and 3.0 Å, yellow between 1.0 and 2.0 Å, and white <1.0 Å movement when the atomic rms are compared. The location of important, flexible Arg residues discussed in the text are labeled for reference.