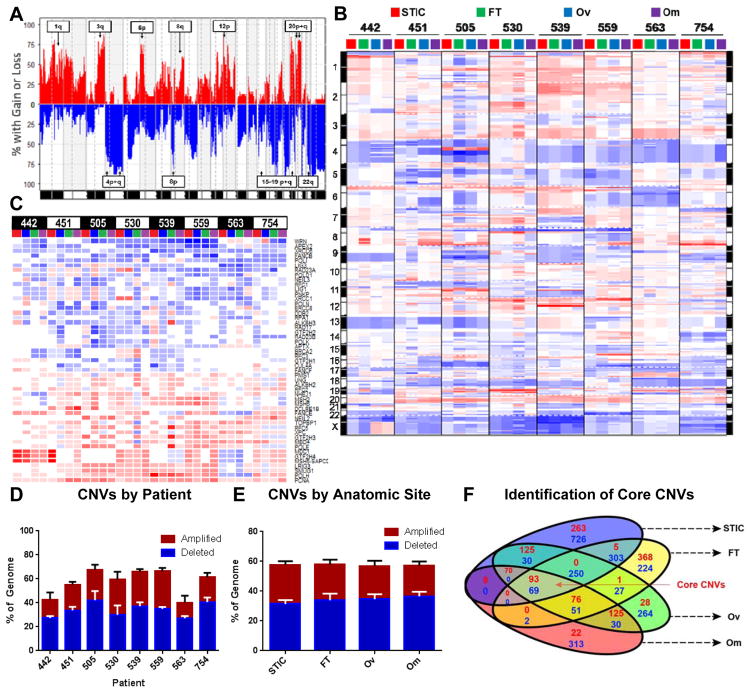
Figure 3. Genomic instability is a core feature of ovarian cancer that frequently involves DNA-damage repair genes.
A) Frequency plot of copy number variations (CNVs) across all patients and all four anatomic sites. Annotated arm level events were also identified as significant in the TCGA analysis of HGSOC. For all plots in Figure 3, amplifications are red and deletions are blue. B) Genomic aberration plot of CNVs across all anatomic sites and patients imply high degree of genomic instability in all anatomic sites. Chromosome numbers are indicated on margin. Amplifications are red; deletions are blue. C) CNV status of significantly altered DNA repair pathway genes (from REPAIRtoire) reveals common deletion of DNA damage response and repair genes known to be involved in HGSOC. Amplifications are red; deletions are blue. D/E) Extent of genomic instability is patient-specific (D) and does not vary by anatomic site (E). F) GISTIC identification and analysis of significantly altered genes identifies conserved core amplifications and deletions. Number of amplifications in red and deletions in blue.