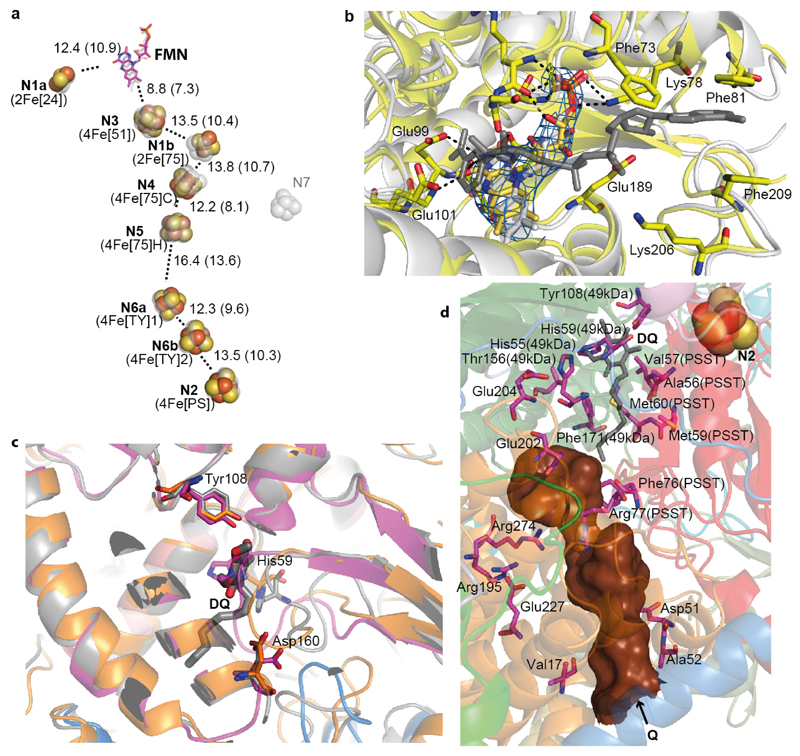
Figure 2. Arrangement of redox centers and substrate binding sites.
a. Fe-S clusters are shown as spheres with centre-to-centre and edge-to-edge (in brackets) distances indicated in Å, overlaid with transparent grey depictions from T. thermophilus. Both traditional and structure-based (in brackets) nomenclature for clusters is shown. b. NADH binding site (overlay with T. thermophilus structure in grey, containing NADH). Cryo-EM density for FMN is shown in blue. Key residues involved in interactions with FMN and NADH are shown as sticks. c. Quinone binding site with subunits coloured as Fig.1. Key β1-β249-kDa loop deviates from bacterial structure (grey) and is more similar to Y. lipolytica (orange, PDB 4WZ712), clashing with the decyl-ubiquinone (DQ) head group position in T. thermophilus (grey). d. Environment surrounding the Q cavity (brown surface, entrance point indicated by an arrow), with some of functionally important residues shown as sticks and labelled with non-ND1 subunit names in brackets. The quinone from the aligned T. thermophilus structure is shown in grey (DQ), demonstrating that the distal part of the cavity is blocked in the ovine enzyme.