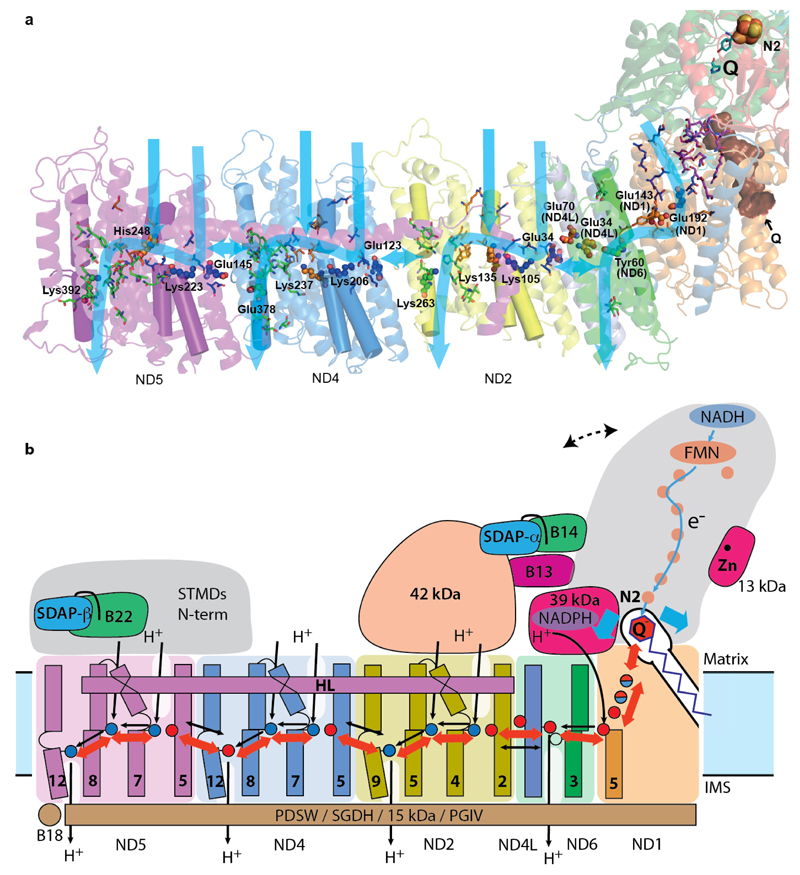
Figure 4. Mechanism of mitochondrial complex I.
a. Structure of the core subunits of ovine complex I coloured as in Fig. 1b, with polar residues in proton channels shown as sticks, with carbon in blue, orange and green for input, connecting and output parts respectively. Key residues GluTM5, LysTM7, Lys/HisTM8 and Lys/GluTM12 from the antiporters and the corresponding residues in the E-channel (near Q site) are shown as small spheres and labelled. These residues sit on flexible loops in discontinuous TM helices shown as cylinders. Polar residues linking the E-channel to the Q cavity (brown) are shown in magenta. Tyr10849-kDa and His5949-kDa are shown in cyan near the position of bound Q in bacteria. Possible proton translocation pathways are indicated by blue arrows. b. Graphic of the coupling mechanism. Core and some putatively regulatory supernumerary subunits are shown. Conformational changes, indicated by red arrows, propagate from the Q site/E-channel to antiporter-like subunits via the central hydrophilic axis. Shifts of helices near the cluster N231 (blue arrows) may help initiate the process. ND5 helix HL and traverse helices from four supernumerary subunits on the IMS side may serve as “stators”. Dashed line indicates the shift of peripheral arm in the “closed” conformation (Extended Data Fig. 8). NADPH-containing 39 kDa subunit and Zn-containing 13 kDa subunit are essential for activity and may serve as redox “sensors”. Both SDAP subunits interact with their LYR partners via “flipped out” phosphopantetheine (black line). The net result of one conformational cycle, driven by NADH:ubiquinone oxidoreduction, is the translocation of four protons across the membrane (black lines indicate possible pathways).