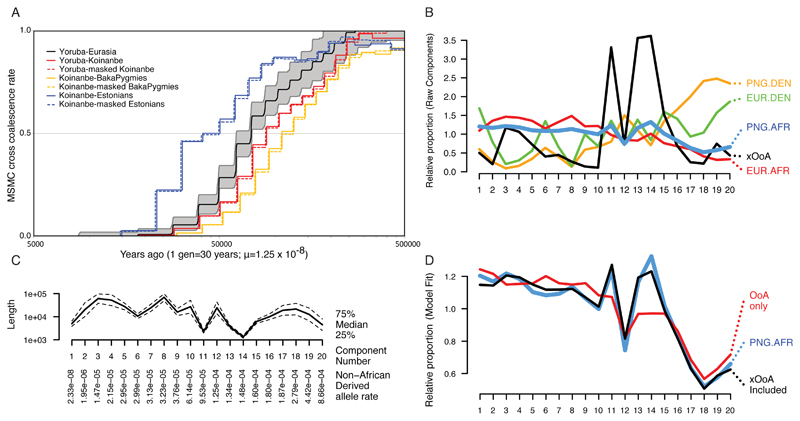
Figure 2. Evidence of an xOoA signature in the genomes of modern Papuans.
Panel A: MSMC split times plot. The Yoruba-Eurasia split curve shows the mean of all Eurasian genomes against one Yoruba genome. The grey area represents top and bottom 5% of runs. We chose a Koinanbe genome as representative of the Sahul populations. Panels B-D: Decomposition of Papuan haplotypes inferred as African by fineSTRUCTURE. Panel B: Semi-parametric decomposition of the joint distribution of haplotype lengths and non-African derived allele rate per SNP, showing the relative proportion of haplotypes in K=20 components of the distribution, ordered by non-African derived allele rate, relative to the overall proportion of haplotypes in each component. The four datasets produced by considering haplotypes inferred as (African/Denisova) in (Europeans/Papuans) are shown with our inferred "extra Out-of-Africa xOoA" component. Panel C: The properties of the components in terms of non-African derived allele rate, on which the components are ordered, and length.
Panel D: The reconstruction of haplotypes inferred as African in the genomes of Papuan individuals, using a mixture of all other data (red) and with the addition of the xOoA signature (black).