ABSTRACT
Insertion sequencing (INSeq) analysis of Rhizobium leguminosarum bv. viciae 3841 (Rlv3841) grown on glucose or succinate at both 21% and 1% O2 was used to understand how O2 concentration alters metabolism. Two transcriptional regulators were required for growth on glucose (pRL120207 [eryD] and RL0547 [phoB]), five were required on succinate (pRL100388, RL1641, RL1642, RL3427, and RL4524 [ecfL]), and three were required on 1% O2 (pRL110072, RL0545 [phoU], and RL4042). A novel toxin-antitoxin system was identified that could be important for generation of new plasmidless rhizobial strains. Rlv3841 appears to use the methylglyoxal pathway alongside the Entner-Doudoroff (ED) pathway and tricarboxylic acid (TCA) cycle for optimal growth on glucose. Surprisingly, the ED pathway was required for growth on succinate, suggesting that sugars made by gluconeogenesis must undergo recycling. Altered amino acid metabolism was specifically needed for growth on glucose, including RL2082 (gatB) and pRL120419 (opaA, encoding omega-amino acid:pyruvate transaminase). Growth on succinate specifically required enzymes of nucleobase synthesis, including ribose-phosphate pyrophosphokinase (RL3468 [prs]) and a cytosine deaminase (pRL90208 [codA]). Succinate growth was particularly dependent on cell surface factors, including the PrsD-PrsE type I secretion system and UDP-galactose production. Only RL2393 (glnB, encoding nitrogen regulatory protein PII) was specifically essential for growth on succinate at 1% O2, conditions similar to those experienced by N2-fixing bacteroids. Glutamate synthesis is constitutively activated in glnB mutants, suggesting that consumption of 2-ketoglutarate may increase flux through the TCA cycle, leading to excess reductant that cannot be reoxidized at 1% O2 and cell death.
IMPORTANCE Rhizobium leguminosarum, a soil bacterium that forms N2-fixing symbioses with several agriculturally important leguminous plants (including pea, vetch, and lentil), has been widely utilized as a model to study Rhizobium-legume symbioses. Insertion sequencing (INSeq) has been used to identify factors needed for its growth on different carbon sources and O2 levels. Identification of these factors is fundamental to a better understanding of the cell physiology and core metabolism of this bacterium, which adapts to a variety of different carbon sources and O2 tensions during growth in soil and N2 fixation in symbiosis with legumes.
KEYWORDS: INSeq, nodules, Rhizobium leguminosarum, legumes, nitrogen fixation, Pisum sativum
INTRODUCTION
Rhizobia are alphaproteobacteria able to form symbioses with legumes, on which they induce development of root nodules. Nodules provide an environment with both a low O2 concentration and the correct nutrients to enable differentiation of rhizobia into nitrogen (N2)-fixing bacteroids (1). Bacteroids fix atmospheric N2 into ammonium (NH4+), which is secreted to the plant host, and are, in turn, provided with carbon that they utilize as an energy source (2). To establish an effective N2-fixing root nodule, a highly specific exchange of signals between plant and bacteria is required. Rhizobia attach to root hairs and grow down plant-derived infection threads into the root cortex (1, 3, 4). During release from infection threads, rhizobia become surrounded by a plant-derived symbiosome membrane, creating a low-O2 environment for N2 fixation to occur (5). Nitrogenase, the enzyme responsible for N2 fixation, requires at least 16 molecules of magnesium-ATP (MgATP) per molecule of N2 fixed (N2+ 8H++ 16MgATP + 8e− → 2NH3+ H2+ 16MgADP + 16Pi) (6). NH3 is not assimilated by bacteroids; instead, it is exported to the plant, where it is incorporated into amino acids which are transported to growing shoots (1).
Symbiotic N2 fixation relies on precise and efficient integration of bacterial and plant metabolism. The plant provides dicarboxylic acids, predominately malate and succinate, and these are taken up by bacteroids via the dicarboxylic transport system (Dct), which is essential for N2 fixation (7–9). Furthermore, either NAD+ malic enzyme (diphosphopyridine nucleotide-dependent malic enzyme [Dme]) or the combined activities of Dme, phosphoenolpyruvate (PEP) carboxykinase, and pyruvate kinase are essential for dicarboxylate metabolism and N2 fixation in bacteroids (9, 10).
Rhizobium leguminosarum bv. viciae strain 3841 (Rlv3841) induces root nodules on viciae legumes, such as the agriculturally important pea (Pisum sativum), vetch (Vicia cracca), and lentil (Lens culinaris) (11, 12). The genome of Rlv3841 is 7.75 Mb, consisting of a circular chromosome (4,788 genes) and six plasmids: pRL12 (790 genes), pRL11 (635 genes), pRL10 (461 genes), pRL9 (313 genes), pRL8 (141 genes), and pRL7 (189 genes) (12). Microarray, transcriptomic, and bioinformatics studies carried out on Rhizobium have indicated many gene functions. However, a high proportion (25%) remain uncharacterized across the genome, and while transcriptional studies allow identification of induced genes, they do not necessarily identify those important for growth in a particular environment (11, 13).
Mariner transposon insertion sequencing (INSeq) is a powerful and robust technique for the study of gene fitness at the genome scale. Libraries of insertion mutants are analyzed by high-throughput sequencing to assess the effect of mutation of a particular gene on growth and survival of the bacterium (14). Mariner transposons insert at thymine-adenine (TA) motifs found abundantly in Rlv3841 (15, 16), and INSeq sequencing data can be coupled with a hidden Markov model (HMM) for analysis (17). The HMM assigns genes to one of four classification states based on the fitness of mutants inferred from their population frequency in the sequenced library: growth essential (cannot tolerate insertion), growth defective (insertion impairs growth), growth neutral (insertion has a neutral impact on growth), or growth advantaged (insertion enhances growth).
C4-dicarboxylates that fuel N2 fixation in the low-O2 environment of bacteroids are catabolized by the O2-dependent tricarboxylic acid (TCA) cycle. Mutational studies of enzymes in the TCA cycle show that it is required for N2 fixation in R. leguminosarum and Sinorhizobium meliloti (18–21), although the entire cycle may not always be essential, as is the case in soybean bacteroids (22, 23). Comparing catabolism of succinate with a sugar, such as glucose, at different O2 tensions, is important in understanding how succinate catabolism is affected by low-O2 environments and how C4-dicarboxylic acids fuel N2 fixation. Here we describe the use of a Rhizobiaceae-compatible MmeI-adapted mariner transposon sequencing vector, pSAM_RI (14), in an INSeq genetic screen of Rlv3841 growth under four different conditions: glucose (10 mM) at 21% O2, succinate (20 mM) at 21% O2, glucose (10 mM) at 1% O2, and succinate (20 mM) at 1% O2. Analysis of the growth requirements for these conditions, particularly growth on succinate at low O2, is relevant to understanding bacteroid metabolism in legume nodules.
RESULTS AND DISCUSSION
Growth of R. leguminosarum and mariner library construction analysis.
The generation times of Rlv3841 were 3.7 h (±0.1) when grown on glucose at 21% O2, 4 h (±0.1) on succinate at 21% O2, 3.8 h (±0.2) on glucose at 1% O2, and 6.2 h (±0.3) on succinate at 1% O2. The combination of succinate and 1% O2 imposed the greatest growth restriction on Rlv3841. This is consistent with succinate being catabolized by the TCA cycle, which generates large amounts of NADH and FADH2 which must then be reoxidized by the electron transport chain. For growth on succinate at 1% O2, electron transport may start to limit growth as reoxidization of the TCA cycle-generated reductant is reliant on the presence of molecular oxygen to act as a final electron acceptor.
From sequencing Rlv3841 INSeq transposon insertion libraries from the four different growth conditions (glucose at 21% O2, succinate at 21% O2, glucose at 1% O2, and succinate at 1% O2), a total of 78 million barcoded sequencing reads were obtained. Sequencing data coupled with HMM analysis enabled the phenotypes of mutants with changes in 7,316 genes to be classified into one of four categories (growth essential, growth defective, growth neutral, or growth advantaged) (see Data set S1 in the supplemental material). No data are available for 21 genes of Rlv3841 which lack TA motifs (14).
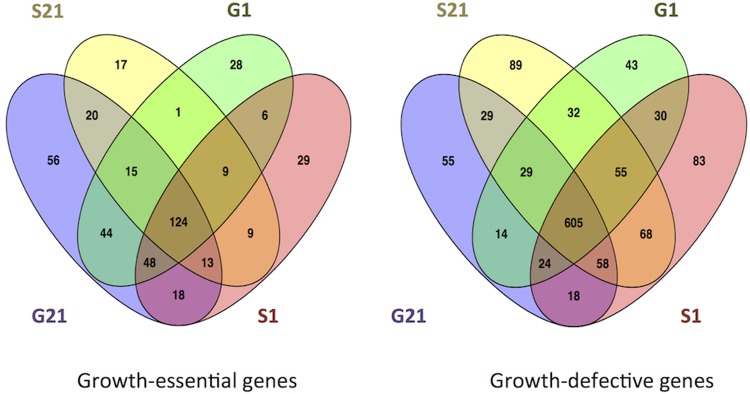
Across the four different growth conditions, mutants of 307 genes (4.2% of the genome) resulted in classification as growth essential, 900 as growth defective (12.3%), 6,072 as (83.3%) growth neutral, and 7 as growth advantaged (0.1%) (Table S3). For 288 genes of unknown function, mutants had a growth-essential, growth-defective, or growth-advantaged phenotype. Under all four conditions, mutants in 124 genes (core genes) were found to be growth essential (Fig. 1). These encode well-characterized essential functions, including translation, transcription, ATP synthesis, membrane transport, and metabolite biosynthesis. A total of 605 genes were identified whose mutants were core growth defective (Fig. 1), encoding functions including transport, protein chaperoning, DNA repair, metabolism, and transcriptional regulation. Included in this category are 127 encoding hypothetical proteins of unknown function. An average of 7 mutants were classified as growth advantaged under each condition (these can be identified in Data set S1), and these genes were distributed on pRL7, pRL9, pRL10, and the chromosome. Mutation of a pseudogene on pRL10 (pRL100096) was growth advantaged specifically on succinate as a carbon source. Pseudogenes are generally considered to be nonfunctional genomic sequences; however, it is possible for them to encode partial activity or effect the mRNA stability of closely related functional genes (24). No mutants were growth advantaged under all growth conditions, consistent with natural selection removing any generally deleterious genes from the population.
FIG 1.
Venn diagram showing number of gene mutants classified as growth essential (ES) and growth defective (GD) under different growth conditions. G21, glucose at 21% O2; S21, succinate at 21% O2; G1, glucose at 1% O2; S1, succinate at 1% O2.
To validate the accuracy of INSeq gene fitness predictions, mutants in five genes, identified by INSeq as growth defective on succinate, were selected for experimental validation. The in vivo mean generation time (MGT) of wild-type Rlv3841 on succinate at 21% O2 (4.0 h) was compared with that of strains mutated in RL0037 (pckA), RL1172, RL3424 (dctA), RL4283 (ptsP), and RL4284 (ask) (Table S1). Each mutant strain had a significantly longer MGT than wild-type Rlv3841 (Table 1). The output data from HMM analysis (number of insertion sites, insertion density, and mean read count) were analyzed for these genes and a fitness value was calculated for each by multiplying insertion density by the number of TA sites and then by the mean read count (Table 1). (Insertion density is the proportion of TA sites containing an insertion in the mutant population, and mean read count is the average number of insertions at TA sites containing at least one insertion.) There was observed to be an inverse relationship between fitness value and MGT (Table 1); the higher the fitness value, the shorter the MGT. Mutation of RL0037 (pckA) led to the lowest fitness value (3.34) and the longest MGT (16.0 h), while mutation of RL1172 gave the highest fitness value (17.29) and shortest MGT (5.0 h). Mutation of RL3424 (dctA) was an exception, having a reasonably high fitness value (13.28) yet one of the longest MGTs (16.0 h). This apparent discrepancy may be explained by most TA motifs in RL3424 being in the gene's termini, so mariner insertion may not have fully disrupted gene function (25).
TABLE 1.
HMM analysis of five INSeq-identified genes, mutation of which leads to a growth-defective phenotype, and mean generation times of mutants on succinate at 21% O2
| Gene | No. of TA sites | Insertion densitya | Mean read countb | Fitness valuec | MGTd (h) |
|---|---|---|---|---|---|
| RL0036 (pckA) | 22 | 0.09 | 1.67 | 3.34 | 16.0 |
| RL1172 | 10 | 0.23 | 7.42 | 17.29 | 5.0 |
| RL3424 (dctA) | 17 | 0.27 | 2.85 | 13.28 | 16.0 |
| RL4283 (ptsP) | 25 | 0.28 | 2.15 | 15.05 | 5.2 |
| RL4284 (ask) | 24 | 0.21 | 1.08 | 5.39 | 6.4 |
Insertion density is defined as the proportion of TA insertion sites containing an insertion in the mutant population relative to the HMM-predicted value.
Mean read count is defined as the mean number of insertion mutants found at insertion sites that contained at least one insertion in the mutant population.
Fitness value is a measure of total gene mutants in the population.
Mean generation time (MGT) is calculated as the time it takes the population to double during exponential growth phase, measured in triplicate.
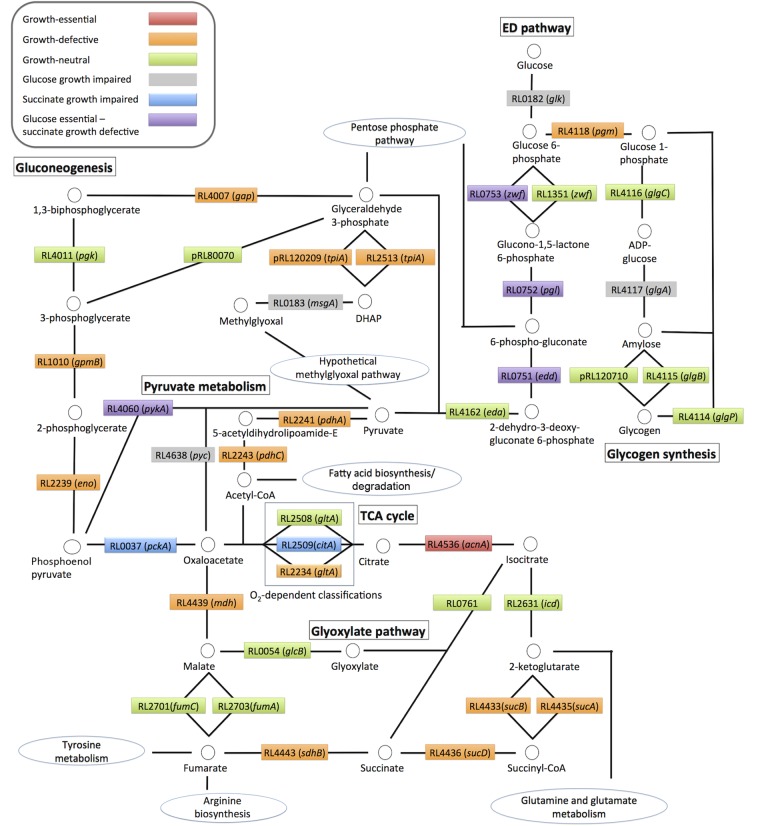
Central metabolism. (i) The ED pathway is needed for optimal growth on both glucose and succinate.
The Entner-Doudoroff (ED) pathway was shown to be essential for growth on glucose, as expected (26), but surprisingly, its mutation caused growth deficiency on succinate (Fig. 2 and Table 2). Most free-living rhizobia utilize the ED pathway for the catabolism of glucose to pyruvate (26, 27), with the pathway converting glucose-6-phosphate into pyruvate in a 1:2 molar ratio and generating one molecule of ATP (27). Its requirement for growth on succinate suggests that sugars made by gluconeogenesis may, at least partly, cycle back through the ED pathway (Fig. 2 and Table 2). Carbon cycling has been reported for a number of other bacterial species, including Sinorhizobium meliloti (26–28). RL0753 and RL1315 encode putative glucose-6-phosphate-1-dehydrogenases (Fig. 2 and Table 2), with these proteins sharing 43% amino acid identity (29). However, the essential classification of RL0753 suggests that it is the “real” zwf with RL1315 unable to compensate. RL0753 was also significantly transcriptionally upregulated on glucose versus succinate (P < 0.05), while expression of RL1315 was unchanged (11). Finally, RL0753 is in an operon with genes encoding enzymes catalyzing subsequent steps in the ED pathway (RL0751 to RL0753). Mutations of all three genes in this cluster are growth essential on glucose and growth defective on succinate (Table 2).
FIG 2.
Central metabolic pathway of Rlv3841 showing the metabolism of glucose and succinate. Candidate genes for enzymes performing the catalytic steps are shown in colored boxes according to their INSeq mutant classification: red, growth essential (ES) under all growth conditions; orange, growth defective (GD) under all growth conditions; green, growth neutral (NE) under all growth conditions; gray, growth impaired on glucose, i.e., growth essential or growth defective specifically on glucose; blue, growth impaired on succinate, i.e., growth essential or growth defective specifically on succinate; purple, growth essential (ES) on glucose and growth defective on succinate.
TABLE 2.
INSeq classification of Rlv3841-encoded enzymes of glucose and succinate central metabolism under different growth conditions
| Locus | Gene | Product description | Pathway(s) | INSeq classificationa for: |
|||
|---|---|---|---|---|---|---|---|
| G21 | S21 | G1 | S1 | ||||
| pRL80070 | Aldehyde dehydrogenase | Gluconeogenesis | NE | NE | NE | NE | |
| pRL120710 | glgB | 1,4-Alpha-glucan branching enzyme | Glycogen synthesis | NE | NE | NE | NE |
| RL0037 | pckA | Phosphoenolpyruvate carboxykinase | Pyruvate metabolism | NE | GD | NE | GD |
| RL0054 | glcB | Malate synthase | Glyoxylate pathway | NE | NE | NE | NE |
| RL0182 | glk | Glucokinase | ED pathway | ES | NE | GD | NE |
| RL0183 | msgA | Methylglyoxal synthase | Hypothetical methyglyoxal pathway | ES | NE | GD | NE |
| RL0751 | edd | Phosphogluconate dehydratase | ED pathway | ES | GD | ES | GD |
| RL0752 | pgl | 6-Phosphogluconolactonase | ED pathway/PPP | ES | GD | ES | GD |
| RL0753 | zwf | Glucose-6-phosphate 1-dehydrogenase | ED pathway/PPP | ES | GD | ES | GD |
| RL0761 | Isocitrate lyase | Glyoxylate pathway | NE | NE | NE | NE | |
| RL1010 | gpmB | Phosphoglycerate mutase | Gluconeogenesis | GD | GD | GD | GD |
| RL1351 | zwf | Glucose-6-phosphate 1-dehydrogenase | ED pathway/PPP | NE | NE | NE | NE |
| RL2234 | gltA | Citrate synthase | TCA cycle | GD | GD | GD | NE |
| RL2239 | eno | Enolase | Gluconeogenesis | GD | GD | GD | GD |
| RL2241 | pdhA | Pyruvate dehydrogenase subunit A | Pyruvate metabolism | GD | GD | GD | GD |
| RL2243 | pdhC | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (PDC) | Pyruvate metabolism | GD | GD | GD | GD |
| RL2508 | gltA | Citrate synthase II | TCA cycle | NE | NE | NE | NE |
| RL2509 | citA | Citrate synthase I | TCA cycle | NE | GD | NE | NE |
| RL2513 | tpiA | Triosephosphate isomerase | Gluconeogenesis/hypothetical methyglyoxal pathway | NE | NE | NE | NE |
| RL2631 | icd | Isocitrate dehydrogenase (NADP) | TCA cycle | NE | NE | NE | NE |
| RL2701 | fumC | Fumarate hydratase class II | TCA cycle | NE | NE | NE | NE |
| RL2703 | fumA | Fumarate hydratase class I, aerobic | TCA cycle | NE | NE | NE | NE |
| RL4007 | gap | Glyceraldehyde-3-phosphate dehydrogenase | Gluconeogenesis | GD | GD | GD | GD |
| RL4011 | pgk | Phosphoglycerate kinase | Gluconeogenesis | NE | NE | NE | NE |
| RL4060 | pykA | Pyruvate kinase | Pyruvate metabolism | ES | GD | ES | GD |
| RL4114 | glgP | Glycogen phosphorylase | Glycogen synthesis | NE | NE | NE | NE |
| RL4115 | glgB | 1,4-Alpha-glucan branching enzyme | Glycogen synthesis | NE | NE | NE | NE |
| RL4116 | glgC | Glucose-1-phosphate adenylyltransferase | Glycogen synthesis | NE | NE | NE | NE |
| RL4117 | glgA | Glycogen synthase | Glycogen synthesis | GD | NE | GD | NE |
| RL4118 | pgm | Phosphoglucomutase | Glycogen synthesis | GD | GD | GD | GD |
| RL4162 | eda | 2-Dehydro-3-deoxyphosphogluconate aldolase | ED pathway | NE | NE | NE | NE |
| RL4433 | sucB | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase | TCA cycle | GD | GD | GD | GD |
| RL4435 | sucA | 2-Oxoglutarate dehydrogenase E1 component | TCA cycle | GD | GD | GD | GD |
| RL4436 | sucD | Succinyl-coenzyme A synthetase alpha chain | TCA cycle | GD | GD | GD | GD |
| RL4439 | mdh | Malate dehydrogenase | TCA cycle | GD | GD | GD | GD |
| RL4443 | sdhB | Succinate dehydrogenase iron-sulfur protein | TCA cycle | GD | GD | GD | GD |
| RL4536 | acnA | Aconitate hydratase | TCA cycle | ES | ES | ES | ES |
| RL4638 | Pyruvate carboxylase | Pyruvate metabolism | GD | NE | GD | NE | |
G21, glucose at 21% O2; S21, succinate at 21% O2; G1, glucose at 1% O2; S1, succinate at 1% O2; ES, growth essential; GD, growth defective; NE, growth neutral.
Intriguingly, mutation of RL4162 (eda), encoding the putative aldolase enzyme catalyzing formation of pyruvate and glyceraldehyde-3-phosphate, resulted in a growth-neutral classification across all conditions (Fig. 2 and Table 3). RL4162 was upregulated on glucose (versus succinate) (11), but its growth-neutral classification suggests that an alternative aldolase is present (29). Sugars are also catabolized by the pentose phosphate pathway (PPP) in Rlv3841 (30). Enzymes catalyzing formation of 6-phospho-gluconate from glucose-6-phosphate were essential, but these reactions are shared by both the ED pathway and the PPP (Fig. 2 and Table 3) (30). There were no essential enzymes unique to the oxidative branch of the PPP.
TABLE 3.
INSeq-identified Rlv3841 genes, mutation of which specifically reduces growth on glucose, together with INSeq classification under different growth conditions
| Locus | Gene | Product description | Classification | INSeq classificationa for: |
|||
|---|---|---|---|---|---|---|---|
| G21 | S21 | G1 | S1 | ||||
| pRL110211 | SBP of ABC transporter, PAAT family | Cell processes | GD | NE | GD | NE | |
| pRL110565 | Transmembrane protein | Cell envelope | ES | NE | ES | NE | |
| pRL120207 | eryD | DeoR family transcriptional regulator (repressor) | Regulation | GD | NE | GD | NE |
| pRL120419 | opaA | Omega-amino acid:pyruvate transaminase | Metabolism of small molecules | GD | NE | GD | NE |
| RL0182 | glk | Glucokinase | Metabolism of small molecules | ES | NE | GD | NE |
| RL0183 | mgsA | Methylglyoxal synthase | Metabolism of small molecules | ES | NE | GD | NE |
| RL0547 | phoB | Phosphate regulon transcriptional regulator PhoB | Regulation | GD | NE | GD | NE |
| RL1479 | Conserved hypothetical protein | Conserved in E. coli | ES | NE | ES | NE | |
| RL1754 | Conserved hypothetical exported protein | Conserved in E. coli | GD | NE | GD | NE | |
| RL2082 | gatB | Aspartyl/glutamyl-tRNAAsn/Gln amidotransferase subunit B | Macromolecule metabolism | ES | NE | GD | NE |
| RL3005 | Hypothetical protein | Unknown function, no known homologues | ES | NE | ES | NE | |
| RL4117 | glgA | Glycogen synthase | Macromolecule metabolism | GD | NE | GD | NE |
| RL4638 | Pyruvate carboxylase | Metabolism of small molecules | GD | NE | GD | NE | |
G21, glucose at 21% O2; S21, succinate at 21% O2; G1, glucose at 1% O2; S1, succinate at 1% O2; ES, growth essential; GD, growth defective; NE, growth neutral.
(ii) The TCA cycle is essential for growth on glucose and succinate.
The chromosome of Rlv3841 contains all the genes for a functional TCA cycle (12), and most genes encoding TCA cycle enzymes showed the same INSeq classification on both glucose and succinate (Fig. 2 and Table 2). Aconitase (encoded by RL4536 [acnA]) was growth essential across all four growth conditions (Fig. 2 and Table 2), consistent with the TCA cycle being essential for growth on both glucose and succinate. Koziol et al. reported that stable Sinorhizobium acnA mutants are possible only in a citrate synthase (gltA) null background, suggesting that intracellular accumulation of citrate is toxic (31). Fumarate hydratase (RL2701 and RL2703) and isocitrate dehydrogenase (RL2631), catalyzing the formation of malate from fumarate and 2-ketoglutarate from isocitrate, respectively, were growth neutral (Table 2). While RL2701 and RL2703 indicate redundancy of fumarate hydratase, there is only a single gene with homology to isocitrate dehydrogenase (RL2631) (29). In both Bradyrhizobium japonicum and S. meliloti, mutation of isocitrate dehydrogenase has no severe effects on growth, and mutants nodulated their respective host plants (32, 33). In S. meliloti, isocitrate dehydrogenase is essential for N2 fixation (32); however, it is not needed in B. japonicum (33). When isocitrate dehydrogenase was mutated (32, 33), it was unknown if and where flux was rerouted.
Three candidate citrate synthase genes are present in Rlv3841: RL2508, RL2509, and RL2234 (12) (Table 2). On succinate at 21% O2, mutation of either RL2234 or RL2509 caused a growth-defective phenotype, while on glucose at both 21% O2 and 1% O2, only RL2234 was required. Optimum growth on a TCA cycle intermediate (succinate) requires greater TCA cycle enzyme activity than does growth on a sugar such as glucose. This may explain why activity of two citrate synthases is needed when growing on succinate, while one is sufficient for growth on glucose. However, at 1% O2 with succinate, mutation of any of the three candidate citrate synthases was growth neutral (Fig. 2 and Table 2). The combination of 1% O2 and succinate results in the longest MGT for Rlv3841 and suggests that TCA cycle activity is severely reduced, minimizing the requirement for citrate synthase isozymes and, presumably, activity (Fig. 2 and Table 2). The TCA cycle generates large amounts of reductant, posing reoxidization problems at such low O2 availability. It is therefore likely that there is reduced flux through the TCA cycle at 1% O2 and bacterial growth is slowed (MGT of 3.7 h on glucose at 1% O2 versus 6.2 h on succinate at 1% O2). Reduced flux through the TCA cycle at low O2 is strikingly similar to metabolism occurring in bacteroids, particularly of soybean, where mutation of genes encoding even crucial TCA cycle enzymes does not prevent N2 fixation (32, 33).
Mutations that specifically reduced growth on glucose at both 1% and 21% O2.
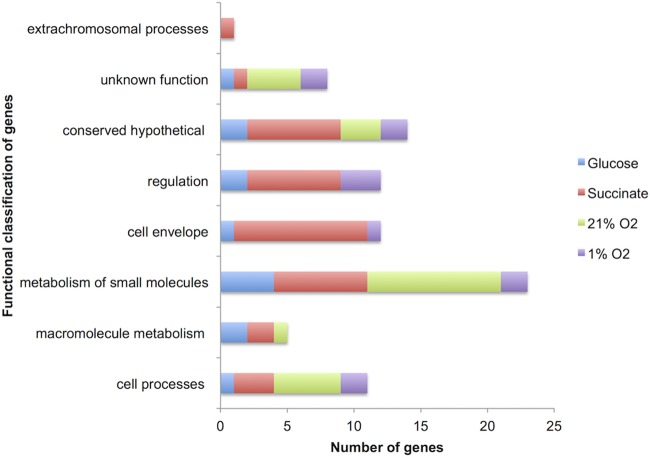
Thirteen genes were required specifically for growth on glucose (i.e., mutation of these genes results in a growth-essential or growth-defective phenotype at both 21% and 1% O2) (Table 3) and encoded functions span regulation, the cell envelope, small-molecule metabolism, macromolecule metabolism, and cell processes (Fig. 3). The largest proportion are involved in small-molecule metabolism (Fig. 3), and a number of these genes have known enzymatic functions in central carbohydrate metabolism; RL0182 (glk) encodes a glucokinase needed for the first step in glucose activation to glucose-6-phosphate (Fig. 2), RL4117 (glgA) encodes a glycogen synthase, and RL4638 encodes a pyruvate carboxylase (Fig. 2). Glycogen synthesis is required for growth on glucose (26), while pyruvate carboxylase has an essential anaplerotic function converting pyruvate to oxaloacetate, for further metabolism in the TCA cycle (34).
FIG 3.
Function distribution of the INSeq-identified genes required specifically for growth on glucose, succinate, 21% O2, and 1% O2 (listed in Tables 3 to 6). Functional classification is based on Riley codes (12).
Growth on glucose required the transcriptional regulators RL0547 (phoB) and pRL120207 (eryD). pRL120207 (eryD) encodes a repressor of the erythritol catabolic operon (35), suggesting that unregulated expression of the erythritol operon interferes with glucose catabolism. The requirement of pRL120207 (eryD) for growth on glucose was not observed in S. meliloti (36, 37). However, this difference highlights how single mutational studies are not necessarily comparable to INSeq, in which mutants are observed in a competition with thousands of other strains. This can allow resolution of growth defects that may not be evident when the mutant is the sole strain present.
RL0547 (phoB) encodes the response regulator of the phosphate limitation (PhoR-PhoB) two-component regulatory system (38). Glucose is metabolized via sugar phosphate intermediates and so phosphate and glucose metabolism are linked, which is not the case for dicarboxylate metabolism. Thus, the requirement for PhoB for growth on glucose suggests that integration with the Pho regulon may be due to the synthesis of phosphorylated intermediates (39, 40).
Rlv3841 required the methylglyoxal pathway alongside the ED pathway for optimal growth on glucose. The methylglyoxal pathway is present in some prokaryotic species, converting dihydroxyacetone phosphate (DHAP) derived from glycolysis or the ED pathway into methylglyoxal and then into pyruvate (41). This pathway is activated by increased concentrations of glucose phosphates in E. coli (42). This suggests that the pathway reduces sugar phosphate concentrations and increases inorganic phosphate without generating ATP (42). We propose that the methylglyoxal pathway may run in parallel to the ED pathway in Rlv3841 to consume excess glucose-6-phosphate and generate pyruvate (Table 4), consequently reducing the concentration of phosphorylated intermediates generated in glucose catabolism. This concept is supported by the clustering of methylglyoxal synthase (RL0183) and glucokinase (RL0182). While mutating methylglyoxal synthase may be polar on glucokinase and impair glucose catabolism, the ED and methylgloxal pathways are clearly linked. A functional methylglyoxal pathway has not previously been identified in rhizobia; however, the committed enzyme in the methylglyoxal pathway, methylglyoxal synthase (RL0183), was required for growth specifically on glucose (Fig. 2 and Table 4). Microarray data show that expression of RL0183 is 2-fold induced on glucose compared to the value for succinate-grown cells (P < 0.01) (13). Bioinformatics analysis indicates that Rlv3841 possesses all necessary genes for a functional methylglyoxal pathway (29, 38) (Table 4) and that the enzymes are well integrated into general central metabolism. Mutations in many of the genes encoding enzymes of this pathway were growth defective on glucose (Table 4). The final two steps of the methylglyoxal pathway have multiple potential candidate genes, which could explain why their mutation is growth neutral (Table 4).
TABLE 4.
Proposed Rlv3841 methylglyoxal metabolic pathway
| Compound | Enzyme (gene[s]) or pathways | INSeq classificationa |
|---|---|---|
| Glucose | ES | |
| ↓ | Glucokinase (RL0182) | |
| Glucose 6-phosphate | ES | |
| ↓ | Pentose phosphate and ED pathways | |
| Glyceraldehyde 3-phosphate | GD | |
| ↓ | Triosephosphate isomerase (pRL120209, RL2513) | |
| Dihydroxyacetone phosphate | GD | |
| ↓ | Methylglyoxal synthase (RL0183) | |
| Methylglyoxal | NE | |
| ↓ | Methylglyoxal reductase (pRL120760, RL3064, RL3243) | |
| Lactate | NE | |
| ↓ | Lactate dehydrogenase (pRL120231, pRL120601, RL0444, RL3578) | |
| Pyruvate |
ES, growth essential; GD, growth defective; NE, growth neutral.
Two further plasmid genes were required for growth on glucose: pRL110565, encoding a putative transmembrane protein, and pRL120419 (opaA), encoding an omega-amino acid:pyruvate transaminase (43). The transmembrane protein possesses a conserved MatE domain, which primarily functions in the export of metabolic and xenobiotic organic cations (29). OpaA synthesizes beta-alanine by the alanine-dependent transamination of pyruvate (44), which is a key intermediate in glucose catabolism. This may help regulate pyruvate levels by buffering with a nonprotogenic amino acid.
Mutations that specifically reduced growth on succinate at both 21% and 1% O2.
C4-dicarboxylates, including succinate, are provided to bacteroids by the plant host during symbiosis to fuel N2 fixation (7). Thirty-eight genes were required specifically for growth on succinate (i.e., mutants are growth essential or growth defective on succinate at both 21% and 1% O2) (Table 5). Functional classification of these genes shows that the largest proportion encodes cell envelope-related proteins (Fig. 3). These included genes encoding proteins within the PrsD-PrsE type I secretion system (RL3657), genes involved in exopolysaccharide (EPS) synthesis (RL4658), and genes encoding transmembrane proteins (RL0423, RL1391, and RL1392). Bioinformatics analysis indicates that RL3654 to RL3657 all have potential functional roles in the cell envelope (29). RL3654 encodes a polysaccharide biosynthesis protein, RL3655 a glycosyltransferase, RL3656 a lipase, and RL3657 (prsE) the permease component of the PrsDE type I secretion system. PrsDE exports the EPS glycanases PlyA and PlyB, which are responsible for correct cleavage of EPS (45, 46). PrsDE is also responsible for export of rhizobial adhesins and other proteins to the outer surface. While mutation of the ATP-binding component of this transporter (RL3658, encoding PrsD) had no significant effect on fitness, microarray data show that RL3658 (prsD) was significantly upregulated on succinate versus glucose (P > 0.05) (11). However, three genes (RL0072, RL0181, and RL0623) encode homologues with 33 to 38% identity to PrsD, suggesting functional redundancy (29, 46).
TABLE 5.
INSeq-identified Rlv3841 genes, mutation of which specifically reduces growth on succinate, together with INSeq classification under different growth conditions
| Locus | Gene | Product description | Classification | INSeq classificationa for: |
|||
|---|---|---|---|---|---|---|---|
| G21 | S21 | G1 | S1 | ||||
| pRL80122 | traA | Conjugal transfer protein TraA | Extrachromosomal element | NE | GD | NE | GD |
| pRL90053 | O-antigen ligase | Cell envelope | NE | GD | NE | GD | |
| pRL90207 | Permease component of ABC transporter, CUT2 family | Cell processes | NE | GD | NE | GD | |
| pRL90208 | codA | Cytosine deaminase | Metabolism of small molecules | NE | GD | NE | GD |
| pRL100388 | LacI family transcriptional regulator (repressor) | Regulation | NE | GD | NE | GD | |
| RL0037 | pckA | Phosphoenolpyruvate carboxykinase | Metabolism of small molecules | NE | GD | NE | GD |
| RL0055 | Conserved hypothetical protein | Conserved in E. coli | NE | ES | NE | ES | |
| RL0423 | Transmembrane protein | Cell envelope | NE | GD | NE | GD | |
| RL0588 | Peptidoglycan binding protein | Cell envelope | NE | GD | NE | GD | |
| RL0754 | ordL | Oxidoreductase | Metabolism of small molecules | NE | GD | NE | GD |
| RL0957 | Conserved hypothetical protein | Conserved in other organism than E. coli | NE | GD | NE | GD | |
| RL1388 | Conserved hypothetical protein | Conserved in other organism than E. coli | NE | GD | NE | GD | |
| RL1390 | Hypothetical protein | Unknown function, no known homologues | NE | GD | NE | GD | |
| RL1391 | Transmembrane protein | Cell envelope | NE | GD | NE | GD | |
| RL1392 | Transmembrane protein | Cell envelope | NE | GD | NE | GD | |
| RL1393 | pbpF | Peptidoglycan biosynthesis/penicillin binding protein | Cell envelope | NE | GD | NE | GD |
| RL1561 | pabC | Aminodeoxychorismate lyase | Metabolism of small molecules | NE | GD | NE | GD |
| RL1641 | MerR family transcriptional regulator | Regulation | NE | GD | NE | GD | |
| RL1642 | Two-component sensor/regulator; transcriptional regulator | Regulation | NE | GD | NE | GD | |
| RL2384 | recG | ATP-dependent DNA helicase | Macromolecule metabolism | NE | ES | NE | GD |
| RL2385 | Conserved hypothetical protein | Conserved in E. coli | NE | ES | NE | GD | |
| RL2478 | Outer membrane protein | Cell envelope | NE | GD | NE | GD | |
| RL2990 | ubiA | para-Hydroxybenzoate-polyprenyltransferase | Metabolism of small molecules | NE | GD | NE | GD |
| RL3424 | dctA | C4-dicarboxylate transport protein | Cell processes | NE | GD | NE | GD |
| RL3425 | dctB | Histidine kinase (C4-dicarboxylate transport) | Regulation | NE | GD | NE | GD |
| RL3426 | dctD | Transcriptional regulator C4-dicarboxylate transport (sigma 54) | Regulation | NE | GD | NE | GD |
| RL3427 | AsnC family transcriptional regulator | Regulation | NE | GD | NE | GD | |
| RL3465 | Conserved hypothetical protein | Conserved in E. coli | NE | GD | NE | GD | |
| RL3468 | prs | Ribose-phosphate pyrophosphokinase | Metabolism of small molecules | NE | ES | NE | ES |
| RL3512 | Conserved hypothetical protein | Conserved in E. coli | NE | GD | NE | GD | |
| RL3654 | Polysaccharide biosynthesis protein | Cell envelope | NE | GD | NE | GD | |
| RL3655 | Glycosyltransferase | Cell envelope | NE | GD | NE | GD | |
| RL3656 | Lipase | Metabolism of small molecules | NE | GD | NE | GD | |
| RL3657 | prsE | Permease component of ABC transporter, export family | Cell processes | NE | GD | NE | GD |
| RL3668 | Serine/threonine protein phosphatase | Macromolecule metabolism | NE | GD | NE | ES | |
| RL4523 | Conserved hypothetical exported protein | Conserved in organisms other than E. coli | NE | GD | NE | ES | |
| RL4524 | ecfL | RNA polymerase ECF sigma factor | Regulation | NE | GD | NE | ES |
| RL4658 | exoB | UDP-glucose 4-epimerase | Cell envelope | NE | GD | NE | ES |
G21, glucose at 21% O2; S21, succinate at 21% O2; G1, glucose at 1% O2; S1, succinate at 1% O2; ES, growth essential; GD, growth defective; NE, growth neutral.
In addition, of the four genes, RL1390 to RL1393, required for growth on succinate, three have predicted functional roles in the cell envelope, with both RL1391 and RL1392 encoding putative transmembrane proteins and RL1393 encoding a putative peptidoglycan biosynthesis protein (Table 5). RL4658 (exoB) encodes UDP-glucose-4-epimerase, required for synthesis of UDP-galactose (47), which is incorporated into the repeating units of EPS and other polysaccharides (48). The high number of cell envelope genes required for growth on succinate suggests that restructuring of the cell surface, involving the PrsD-PrsE type I secretion system and UDP-galactose production in particular, is required to enable growth on organic acids.
Genes encoding five transcriptional regulators were required for growth on succinate: pRL100388, RL1641, RL1642, RL3427, and RL4524 (ecfL) (Table 5). pRL100388 encodes a putative LacI family transcriptional regulator with homology to a gluconate utilization system transcriptional repressor found in many prokaryotes (29, 49). RL1641 and RL1642 encode two-component sensor/regulator units belonging to the MerR family of transcriptional regulators. RL3427 encodes an AsnC family transcriptional regulator, which could possibly be involved in regulation of genes encoding the dicarboxylic acid transport (Dct) system located adjacently. As expected, mutants with changes in the dicarboxylic acid transport system (dctABD) (RL3424 to RL3426) were growth defective on succinate (Table 5). Mutations in the dct system have been shown to impair uptake of succinate into cells, leading to growth arrest (50–53). RL4524 (ecfL) encodes an extracytoplasmic function (ECF) sigma factor (54). ECF sigma factors regulate transcription in response to extracellular stimuli (55). Their activity is commonly under the regulation of an anti-sigma factor (ASF), but RL4524 is one of only four Rlv3841 ECF genes not adjacent to an ASF gene. RL4524 expression increased in the presence of pea root exudate, presumably due to an unknown compound (54), and was significantly upregulated in the pea rhizosphere (versus Rlv3841 grown on glucose and ammonia) (13). From results shown here, it may be that EcfL (RL4524) responds to C4-dicarboxylic acids present in pea root exudates.
Mutations of two genes involved in nucleobase synthesis, pRL90208 and RL3468, were growth deficient and growth essential, respectively, on succinate. pRL90208 (codA) encodes a putative cytosine deaminase that functions in pyrimidine metabolism (38). Both CodA and the protein product of RL0891 are suggested to possess deaminase activity utilized in conversion of cytosine to uracil and of 5-methylcytosine to thymine (38). INSeq analysis showed RL0891 to be to be nonessential, suggesting that its product is unable to compensate for the activity of CodA, whose gene insertion mutation was growth defective on succinate. RL3468 (prs) encodes a ribose-phosphate pyrophosphokinase catalyzing biosynthesis of 5-phosphoribosyl diphosphate (PRPP) from ribose 5-phosphate. PRPP is utilized in both purine and pyrimidine biosynthesis, with the first committed step to purine biosynthesis utilizing PRPP as a substrate (56). This suggests that for optimal growth on succinate, there is a requirement for increased nucleotide recovery or synthesis compared to growth on glucose. RL0037 (pckA) encodes phosphoenolpyruvate carboxykinase, which converts oxaloacetate into phosphoenolpyruvate, essential for gluconeogenesis (Fig. 2) (9). RL2990 (ubiA) encodes a 4-hydroxybenzoate octaprenyltransferase required for ubiquinone biosynthesis and electron transport. Cells growing on a TCA cycle intermediate are likely to have a greater requirement for reoxidation of NADH/FADH2 by the electron transport chain. RL1561 (pabC) was also found to be required, and the gene encodes an enzyme in the shikimate pathway also needed for ubiquinone synthesis. This agrees with a greater need for ubiquinone in succinate-grown cells.
Mutations that reduced growth at 21% O2 on both glucose and succinate.
Twenty-three genes were required for optimal growth at 21% O2 on both carbon sources (i.e., mutants are growth essential or growth defective on both glucose and succinate at 21% O2) (Table 6) and encoded functions spanning small-molecule metabolism, macromolecule metabolism, and cell processes (Fig. 3). The largest proportion is involved in small-molecule metabolism (Fig. 3), and 11 genes are predicted to encode enzymes utilizing oxygen:cytochrome c oxidase subunits (RL1021 and RL1022), a deoxygenase (RL0802), a hydroperoxide resistance protein (RL2927), a peroxiredoxin (RL2440), three oxidoreductases (RL0847 [an IMP oxidoreductase], RL3834 [an ErfK oxidoreductase], and RL4186), a mitochondrial respiratory chain complex assembly factor (RL0920), and two adenylate cyclases (RL2441 and RL2926). None of the proteins encoded by these genes are required at 1% O2, presumably because their function is to protect against oxygen toxicity.
TABLE 6.
INSeq-identified Rlv3841 genes, mutation of which specifically reduces growth at 21% O2, together with INSeq classification under different growth conditions
| Locus | Gene | Product description | Classification | INSeq classificationa for: |
|||
|---|---|---|---|---|---|---|---|
| G21 | S21 | G1 | S1 | ||||
| pRL90042 | Pseudogene, conserved hypothetical protein | Not classified (cryptic gene) | GD | GD | NE | NE | |
| pRL120796 | Hypothetical exported protein | Unknown function, no known homologues | GD | GD | NE | NE | |
| RL0802 | Deoxygenase | Metabolism of small molecules | GD | GD | NE | NE | |
| RL0847 | guaB | IMP dehydrogenase | Metabolism of small molecules | ES | ES | NE | NE |
| RL0920 | ATP-binding Mrp family protein | Metabolism of small molecules | GD | GD | NE | NE | |
| RL0921 | Cationic transport protein, CorA family | Cell processes | GD | GD | NE | NE | |
| RL0922 | kup | Potassium uptake transport system protein | Cell processes | GD | GD | NE | NE |
| RL1021 | coxB | Cytochrome c oxidase polypeptide II precursor (cytochrome aa3 subunit 2) | Metabolism of small molecules | GD | GD | NE | NE |
| RL1022 | coxA | Cytochrome c oxidase polypeptide I (cytochrome aa3 subunit 1) | Metabolism of small molecules | GD | GD | NE | NE |
| RL1547 | cvpA | Colicin V production protein | Cell processes | GD | GD | NE | NE |
| RL1548 | radA | DNA repair protein RadA homologue | Macromolecule metabolism | GD | GD | NE | NE |
| RL2440 | Peroxiredoxin | Metabolism of small molecules | GD | GD | NE | NE | |
| RL2441 | Adenylate cyclase | Metabolism of small molecules | GD | GD | NE | NE | |
| RL2926 | Adenylyl cyclase | Metabolism of small molecules | GD | GD | NE | NE | |
| RL2927 | ohrB | Organic hydroperoxide resistance protein | Cell processes | GD | GD | NE | NE |
| RL3180 | Hypothetical protein | Unknown function, no known homologues | GD | GD | NE | NE | |
| RL3181 | Conserved hypothetical protein | Conserved in organisms other than E. coli | GD | GD | NE | NE | |
| RL3182 | Conserved hypothetical protein | Conserved in organisms other than E. coli | GD | GD | NE | NE | |
| RL3507 | Phosphoesterase | Not classified (included assignments) | ES | GD | NE | NE | |
| RL3596 | Conserved hypothetical protein | Conserved in organisms other than E. coli | GD | GD | NE | NE | |
| RL3597 | DEAD box ATP-dependent RNA helicase protein | Cell processes | GD | GD | NE | NE | |
| RL3834 | ErfK/YbiS/YhnG oxidoreductase | Metabolism of small molecules | GD | GD | NE | NE | |
| RL4186 | Oxidoreductase | Metabolism of small molecules | GD | GD | NE | NE | |
G21, glucose at 21% O2; S21, succinate at 21% O2; G1, glucose at 1% O2; S1, succinate at 1% O2; ES, growth essential; GD, growth defective; NE, growth neutral.
Other genes required for optimal growth on 21% O2 include RL0921, predicted to encode a CorA magnesium transporter, and RL0922, encoding a potassium uptake protein (29). Cationic transporters are electrochemical potential driven, and activity is dependent on the oxidation state of the metal ion species (57). Insertion mutation of these enzymes did not impair growth at 1% O2, perhaps because transport of ions in their more reduced state does not limit growth or because other transporters play a greater role at low O2. Rlv3841 possesses an MgtE magnesium transporter channel (RL1461 [mgtE]) that is essential for growth when both the magnesium concentration is limiting and the pH is low, which is also essential for N2 fixation on specific legumes (58). CorA may be the primary transporter of magnesium at 21% O2 and another transporter, such as MgtE, may play a greater role at decreased O2.
RL3597 encodes a putative RNA helicase protein, involved in mRNA biogenesis, and RL1548 (radA) encodes a putative DNA repair protein (29, 59). The observation that their mutants are growth defective is presumably due to their preventative role in oxidative damage to nucleic acids.
Mutations that reduced growth at 1% O2 on both glucose and succinate.
Twelve genes were required for optimal growth at 1% O2 on both succinate and glucose (i.e., mutants are growth essential or growth defective on both glucose and succinate at 1% O2) (Table 7) and encoded functions spanning regulation, the cell envelope, small-molecule metabolism, and cell processes (Fig. 3). The largest proportion is involved in regulation, and three potential low-O2 growth regulators were identified: pRL110072, RL0546 (phoU), and RL4042. pRL110072 encodes a putative GntR family transcriptional regulator, and RL4042 encodes a putative AsrR family transcriptional regulator.
TABLE 7.
INSeq-identified Rlv3841 genes, mutation of which specifically reduces growth at 1% O2, together with INSeq classification under different growth conditions
| Locus | Gene | Product description | Classification | INSeq classificationa for: |
|||
|---|---|---|---|---|---|---|---|
| G21 | S21 | G1 | S1 | ||||
| pRL80039 | Pseudogene | Not classified | NE | NE | GD | GD | |
| pRL100141 | Pseudogene, conserved hypothetical protein | Not classified | NE | NE | GD | GD | |
| pRL110016 | Conserved hypothetical protein | Conserved in organisms other than E. coli | NE | NE | GD | GD | |
| pRL110072 | GntR family transcriptional regulator | Regulation | NE | NE | GD | GD | |
| RL0546 | phoU | Phosphate uptake regulator PhoU | Regulation | NE | NE | GD | GD |
| RL1506 | relA | Stringent response protein | Cell processes | NE | NE | GD | GD |
| RL2585 | nodX | Nodulation protein NodX (probable sugar acetylase) | Metabolism of small molecules | NE | NE | GD | GD |
| RL3412 | Conserved hypothetical protein | Conserved in E. coli | NE | NE | GD | GD | |
| RL4042 | AsrR family transcriptional regulator | Regulation | NE | NE | GD | GD | |
| RL4693 | Polysaccharide deacetylase | Cell envelope | NE | NE | GD | GD | |
| RL4694 | (Di)nucleoside polyphosphate hydrolase | Metabolism of small molecules | NE | NE | GD | GD | |
| RL4695 | bfr | Bacterioferritin | Cell processes | NE | NE | GD | GD |
G21, glucose at 21% O2; S21, succinate at 21% O2; G1, glucose at 1% O2; S1, succinate at 1% O2; ES, growth essential; GD, growth defective; NE, growth neutral.
RL0546 (phoU) is annotated to encode a putative phosphate transport regulatory protein. While the role of PhoU in R. leguminosarum has not been investigated, it has been well studied for E. coli, in which the protein forms part of the phosphate-sensing pathway that negatively regulates the Pho regulon (60). Mutants deficient in phoU show constitutive expression of Pho regulon genes (60–62), encoding proteins involved in phosphate metabolism, energy production, membrane transport, flagellum synthesis, and transcription factors, including a stringent response protein (63). However, it is not known how similar the mechanisms of PhoU regulation are between E. coli and Rlv3841. PhoU may be involved in growth regulation under low O2 through a role in the coordination of phosphate and energy metabolism. The generation of ATP is dependent on the transport of phosphate in the cell, and energy metabolism is O2 dependent.
While RL2585 is annotated as NodX, the presence of which enables a few strains of Rhizobium to nodulate Afghanistan peas (29), the protein encoded by RL2585 shows no significant identity to the characterized NodX (UniProtKB accession number P08888) (29). However, RL2585 possesses a conserved acyltransferase domain, and that might be required for modification of a surface-exposed sugar. RL4693 encodes a putative polysaccharide deacetylase protein, RL4694 a putative dinucleoside polyphosphate hydrolase, and RL4695 a putative bacterioferritin. RL1506 encodes RelA, a central stringent response protein controlling the synthesis of (p)ppGpp, required for adaptation to stress conditions. RelA coordinates responses to the abundance of nitrogen and carbon in Rhizobium (64). In addition to these roles, RelA is suggested to regulate the stress response to oxygen limitation in Bacillus subtilis (65), with a relA deletion mutant strain unable to synthesize the heat shock proteins induced in response to oxygen limitation in the wild type (65). The exact mechanism of this regulation is unclear and does not appear to have been studied for any other bacterial species. However, the requirement of RL1506 (relA) for growth specifically at low oxygen suggests that RelA also regulates a stress response resulting from oxygen limitation in Rlv3841.
Mutations that specifically reduced growth on succinate at 1% O2.
There were a number of mutations resulting in a growth-essential or growth-defective phenotypes specific to one condition but growth neutral across all other conditions. Fifty-two mutations were specific to glucose at 21% O2, 41 specific to succinate at 21% O2, 29 specific to glucose at 1% O2, and 65 specific to succinate at 1% O2. The combination of 1% O2 and succinate is most relevant to symbiotic conditions, with 64 genes classified as causing a growth-defective phenotype upon insertion mutation and a single growth-essential gene, RL2393 (glnB). The MGT of Rlv3841 on succinate at 1% O2 was almost double that of the other three growth conditions (6.2 h compared to 3.7 h, 3.8 h, and 4 h). Under stress, Rlv3841 may be particularly susceptible to mutations altering growth, possibly explaining the higher number of mutations that specifically effected growth under this condition.
The sole gene identified as growth essential on succinate at 1% O2, RL2393 (glnB), encodes the nitrogen regulatory protein PII. This protein forms part of the two-component signaling system that regulates nitrogen-responsive proteins, including glutamine synthetase. Null mutations in glnB cause constitutive activation of ammonia assimilation in R. leguminosarum, promoting increased synthesis of glutamate and glutamine (66, 67). It is intriguing that only on succinate at 1% O2, conditions similar to that experienced by bacteroids, is the activity of RL2393 (glnB) essential. GlnB is downregulated during bacteroid differentiation (68), and microarray data show that expression of glnB is increasingly downregulated from 7 to 28 days in pea bacteroids, suggesting that GlnB is required only prior to symbiotic N2 fixation (13). Constitutive activation of nitrogen assimilation in an RL2393 (glnB) mutant may cause too much 2-ketoglutarate, used for glutamate synthesis, to be removed from the TCA cycle. Depletion of 2-ketoglutarate would force a turn of the TCA cycle to replace it and lead to the production of extra reductant that must then be deoxidized. In a 1% O2 environment, reoxidation of NADH via the electron transport chain may be limited by the O2 available.
A plasmid-specific toxin-antitoxin system on pRL10.
Mutations to genes in the putative operon pRL100005-10 were growth essential or growth defective across all four conditions, and the operon is predicted to encode a putative toxin-antitoxin system (29). The operon consists of two candidate toxin genes (pRL100005 and pRL00009) and three candidate antitoxin or transcriptional regulator component genes (pRL00006, pRL00007, and pRL100010) (see Data set S1) (29). The two candidate toxin genes encode PIN domain proteins, which commonly confer toxicity due to RNase activity and require coexpression of antitoxin components to ensure cell viability (69). Finding a novel toxin-antitoxin system on pRL10 accords with the ability to cure Rlv3841 of each plasmids, with the exception of pRL10 (70). Consideration of this novel toxin-antitoxin system is important if trying to generate new plasmidless rhizobial strains.
Conclusion.
In summary, an INSeq screen of Rlv3841 grown on glucose and succinate at both 21% and 1% O2 enabled identification of factors needed for growth on different carbon sources and O2 levels. Use of HMM analysis assigned gene phenotype classifications to 7,316 genes in the genome of Rlv3841 across four different growth conditions. Analysis of central carbon metabolism pathways allowed roles in glucose and succinate metabolism to be distinguished and the ability of pathways to compensate for single enzyme mutations to be phenotypically quantified. A hypothetical methylglyoxal pathway is proposed for Rlv3841, and enzymes of this pathway were found to be required for growth on glucose. One of the most dramatic observations is that only mutation of RL2393 (glnB) caused a growth-essential phenotype on succinate at 1% O2. Since glnB mutants of R. leguminosarum are constitutively activated for ammonia assimilation, this suggests that removal of 2-ketoglutarate from the TCA cycle by glutamate synthesis is highly deleterious when C4-dicarboxylates are the carbon source in a low-O2 environment, conditions mimicking those of N2-fixing bacteroids in symbiosis with legumes.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Rlv3841 strains listed in Table S1 in the supplemental material were grown at 28°C in tryptone-yeast extract (TY) (71) or universal minimal salts (UMS) medium with appropriate carbon and nitrogen sources. UMS medium is derived from AMS (72), with the alterations made being CaCl2·2H2O (0.51 mM), CoCl2·6H2O (4.2 μM), EDTA-Na2 (1 μM), and FeSO4·7H2O (0.04 mM). All Escherichia coli strains (Table S1) were grown at 37°C in Lennox (L) broth or on L agar (73). Antibiotics were used at the following concentrations (in micrograms per milliliter) unless otherwise stated: gentamicin, 20 (10 for E. coli); kanamycin, 20; neomycin, 40; spectinomycin, 50; streptomycin, 500; and tetracycline, 2 in UMS and 5 in TY. The donor E. coli strain, SM10λpir carrying the mariner INSeq vector pSAM_Rl, was supplemented with 50 μg/ml of neomycin to select for the plasmid and 100 μg/ml of ampicillin (Amp) to select for E. coli.
Generation time of Rlv3841.
To assess growth, 2 × 107 Rlv3841 cells were grown in 500 ml of UMS containing 100 mM ammonium chloride and 500 μl of 1,000× stock vitamins. Either glucose (10 mM) or succinate (20 mM) was added as a carbon source. The cultures were grown at 25°C under either atmospheric conditions (21% O2) or at 1% O2 in an oxygen cabinet (Belle Technology). For cultures (biological triplicates), optical density (at 600 nm [OD600]) measurements were taken at 4-h intervals until growth reached stationary phase. The mean generation time (MGT) of Rlv3841 was calculated as the number of hours it took the population to double while in exponential growth phase. To assess in vivo growth of growth-defective mutants, wild-type Rlv3841 and pK19-generated gene mutants (listed in Table S1) were grown on TY agar slopes. Growth was measured at 21% O2 with succinate (20 mM) as used for mutant library DNA extractions, and OD600 measurements at 4-h intervals were taken in triplicate.
Mariner library construction.
Donor mariner transposon E. coli cells were grown in L broth culture overnight, and recipient rhizobial cells were grown on a TY agar slope. Cultures were subjected to three cycles of pelleting at 6,000 rpm for 5 min, followed by resuspension in TY medium. Donor and recipient cells were pooled in equal ratios, pelleted at 6,000 rpm for 5 min, and resuspended in 30 μl of TY medium. Cell suspensions were spotted onto nitrocellulose filters on TY agar plates and incubated at 27°C overnight, and then the bacteria from the filters were resuspended in UMS medium plus 15% glycerol. Enumeration of total transposon insertion Rlv3841 was performed on 500 μg/ml of streptomycin plus 50 μg/ml of neomycin TY agar. Independent pools of transposon mutants were generated by plating three libraries on 500 μg/ml of streptomycin plus 50 μg/ml of neomycin TY agar and pooling of rhizobial mutants.
Cultures were prepared as described above to measure MGT, with the substitution of 2 × 107 cells from the mariner transposon mutant library. At mid-exponential growth (OD600 of 0.5), DNA was extracted from approximately 109 cells using a Qiagen DNeasy blood and tissue kit for Gram-negative bacteria according to the manufacturer's protocol, with the following modifications: 2 μl of RNase A (100 mg/ml) was added in parallel to proteinase K, and DNA was eluted in 100 μl of Milli-Q water. DNA extraction for three biological replicates generated 12 different library pools. To confirm successful rhizobial DNA extraction, samples were run under electrophoresis on an agarose gel alongside a GeneRuler 1-kb DNA ladder (Thermo Scientific). DNA was quantified with a NanoDrop spectrophotometer (Thermo Scientific).
Library preparation and sequencing.
Three independent pools of 2 × 107 transposon mutants were grown for 14 generations under four different growth conditions: glucose at 21% O2, succinate at 21% O2, glucose at 1% O2, and succinate at 1% O2. Library preparation was carried out independently for each of the mutant libraries. Transposon tags were prepared for DNA sequencing using a modified version of the INSeq method to make the process compatible with the Ion Proton system sequencing platform (74). Linear PCR products were amplified using Ion Proton-compatible BioSAM primers (14) with an annealing temperature of 58.6°C and 500 ng of template DNA. Primer and adaptor sequences are detailed in Table S2. Linear PCR products were purified (GeneJET PCR purification kit) and eluted in 50 μl of Milli-Q water. The biotinylated linear PCR products were bound for affinity capture with Pierce streptavidin magnetic beads (Thermo Scientific), and the enzymatic library preparation steps were performed with the substitution of Klenow (New England BioLabs), Random Primer 6 (New England BioLabs), and T4 DNA ligase (New England BioLabs). A custom INSeq library adapter was used in the adapter ligation step, and then final PCR amplification used sequencing template-incorporated fusion primers designed to be compatible with Ion amplicon library preparation. The 12 INSeq libraries were barcoded using 12 different forward fusion primers which gave different IonXpress barcode sequences for downstream sequence separation. An Ion Proton system reverse-sequencing target, trP1, was used in conjunction with these forward barcode primers. This final sequencing template was 187 bp in length and was gel purified from the PCR products with a GeneJET gel purification kit (Thermo Scientific). DNA to be used for sequencing was analyzed with a bioanalyzer high-sensitivity DNA chip (Agilent Technologies) to ensure its quality and molarity.
Libraries were diluted to 100 pM and pooled. The libraries (100 pM DNA in 25 μl) underwent automated template preparation using an Ion Chef (Life Technologies). DNA sequencing was performed on an Ion Proton system sequencer.
Transposon insertion analysis with a four-state HMM.
Sequencing reads were uploaded onto a Linux server for analysis as previously described (14). In short, “cutadapt” (75) was used for quality trimming and removal of adapter sequences, and the resulting 15- to 16-bp tn-tags were checked for a leading TA motif with a custom-written Perl script. These tags were mapped to the Rlv3841 reference genome using the Bowtie short-read aligner (76) and grouped into Rlv3841 replicons. The files were converted to a .wig format by a custom-written Perl script for further analysis using the Tn-HMM Python module (17). This first calculates the HMM state of each individual TA site and then determines the state of all TA sites within gene boundaries to assign each gene a state as a whole. HMM analysis assigned genes to one of four classification states: growth essential, growth defective, growth neutral, or growth advantaged. The Tn-HMM Python module was used to quantify the number of genes assigned to each state. Gene annotations were from RlegDB (http://rlegdb.jic.ac.uk/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Environment Research Council (grant number NE/L501530/1) and the Biotechnology and Biological Sciences Research Council (grant numbers BB/K001868/1 and BB/K001868/2).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00572-16.
REFERENCES
- 1.Udvardi MK, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 2.Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ, Poole PS. 2009. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc Natl Acad Sci U S A 106:12477–12482. doi: 10.1073/pnas.0903653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terpolilli JJ, Hood GA, Poole PS. 2012. What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv Microb Physiol 60:326. [DOI] [PubMed] [Google Scholar]
- 4.Oldroyd GE, Downie JA. 2004. Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5:566–576. doi: 10.1038/nrm1424. [DOI] [PubMed] [Google Scholar]
- 5.Day DA, Kaiser BN, Thomson R, Udvardi MK, Moreau S, Puppo A. 2001. Nutrient transport across symbiotic membranes from legume nodules. Funct Plant Biol 28:669–676. doi: 10.1071/PP01028. [DOI] [Google Scholar]
- 6.Lodwig EM, Poole PS. 2003. Metabolism of Rhizobium bacteroids. CRC Crit Rev Plant Sci 22:37–78. doi: 10.1080/713610850. [DOI] [Google Scholar]
- 7.Lodwig EM, Hosie AH, Bourdès A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS. 2003. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 8.White J, Prell J, James EK, Poole PS. 2007. Nutrient sharing between symbionts. J Plant Physiol 144:604–614. doi: 10.1104/pp.107.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulley G, Lopez-Gomez M, Zhang Y, Terpolilli J, Prell J, Finan T, Poole PS. 2010. Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation. J Bacteriol 192:4944–4953. doi: 10.1128/JB.00294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Aono T, Poole PS, Finan TM. 2012. NAD(P)+-malic enzyme mutants of Sinorhizobium sp. strain NGR234, but not Azorhizobium caulinodans ORS571, maintain symbiotic N2 fixation capabilities. J Appl Environ Microbiol 78:2803–2812. doi: 10.1128/AEM.06412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karunakaran R, Ramachandran VK, Seaman JC, East AK, Mouhsine B, Mauchline TH, Prell J, Skeffington A, Poole PS. 2009. Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J Bacteriol 191:4002–4014. doi: 10.1128/JB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young JPW, Crossman LC, Johnston AW, Thomson NR, Ghazoui ZF, Hull KH, Parkhill J. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramachandran VK, East AK, Karunakaran R, Downie JA, Poole PS. 2011. Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12:R106. doi: 10.1186/gb-2011-12-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry BJ, Yost CK. 2014. Construction of a mariner-based transposon vector for use in insertion sequence mutagenesis in selected members of the Rhizobiaceae. BMC Microbiol 14:298. doi: 10.1186/s12866-014-0298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picardeau M. 2010. Transposition of fly mariner elements into bacteria as a genetic tool for mutagenesis. Genetica 138:551–558. doi: 10.1007/s10709-009-9408-5. [DOI] [PubMed] [Google Scholar]
- 16.Plasterk RH, Izsvák Z, Ivics Z. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet 15:326–332. doi: 10.1016/S0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 17.DeJesus MA, Ioerger TR. 2013. A hidden Markov model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14:303. doi: 10.1186/1471-2105-14-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walshaw DL, Wilkinson A, Mundy M, Smith M, Poole PS. 1997. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology 143:2209–2221. doi: 10.1099/00221287-143-7-2209. [DOI] [PubMed] [Google Scholar]
- 19.Dunn MF. 1998. Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia. FEMS Microbiol Rev 22:105–123. doi: 10.1111/j.1574-6976.1998.tb00363.x. [DOI] [PubMed] [Google Scholar]
- 20.Driscoll BT, Finan TM. 1993. NAD+-dependent malic enzyme of Rhizobium meliloti is required for symbiotic nitrogen fixation. Mol Microbiol 7:865–873. doi: 10.1111/j.1365-2958.1993.tb01177.x. [DOI] [PubMed] [Google Scholar]
- 21.Gardiol A, Arias A, Cervenansky C, Martinez-Drets G. 1982. Succinate dehydrogenase mutant of Rhizobium meliloti. J Bacteriol 151:1621–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green LS, Emerich DW. 1997. The formation of nitrogen-fixing bacteroids is delayed but not abolished in soybean infected by an [alpha]-ketoglutarate dehydrogenase-deficient mutant of Bradyrhizobium japonicum. J Plant Physiol 114:1359–1368. doi: 10.1104/pp.114.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green LS, Emerich DW. 1999. Is bacteroid α-ketoglutarate dehydrogenase needed for nitrogen fixation? p 37–40. In Mart́inez E, Herńandez G (ed), Highlights of nitrogen fixation research. Springer US, New York, NY. [Google Scholar]
- 24.Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ, Harte R, Balasubramanian S, Tanzer A, Diekhans M, Reymond A. 2012. The GENCODE pseudogene resource. Genome Biol 13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geddes BA, Oresnik IJ. 2014. Physiology, genetics, and biochemistry of carbon metabolism in the alphaproteobacterium Sinorhizobium meliloti. Can J Microbiol 60:491–507. doi: 10.1139/cjm-2014-0306. [DOI] [PubMed] [Google Scholar]
- 27.Fuhrer T, Fischer E, Sauer U. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187:1581–1590. doi: 10.1128/JB.187.5.1581-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portais JC, Tavernier P, Gosselin I, Barbotin JN. 1999. Cyclic organization of the carbohydrate metabolism in Sinorhizobium meliloti. Eur J Biochem 265:473–480. doi: 10.1046/j.1432-1327.1999.00778.x. [DOI] [PubMed] [Google Scholar]
- 29.Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat Genet 3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 30.McKay IA, Dilworth MJ, Glenn AR. 1989. Carbon catabolism in continuous cultures and bacteroids of Rhizobium leguminosarum MNF 3841. Arch Microbiol 152:606–610. doi: 10.1007/BF00425495. [DOI] [Google Scholar]
- 31.Koziol U, Hannibal L, Rodríguez MC, Fabiano E, Kahn ML, Noya F. 2009. Deletion of citrate synthase restores growth of Sinorhizobium meliloti 1021 aconitase mutants. J Bacteriol 191:7581–7586. doi: 10.1128/JB.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDermott TR, Kahn ML. 1992. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J Bacteriol 174:4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah R, Emerich DW. 2006. Isocitrate dehydrogenase of Bradyrhizobium japonicum is not required for symbiotic nitrogen fixation with soybean. J Bacteriol 188:7600–7608. doi: 10.1128/JB.00671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lodiwg EM. 2001. Regulation of carbon metabolism in Rhizobium leguminosarum. PhD thesis. University of Reading, Reading, United Kingdom. [Google Scholar]
- 35.Yost CK, Rath AM, Noel TC, Hynes MF. 2006. Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 152:2061–2074. doi: 10.1099/mic.0.28938-0. [DOI] [PubMed] [Google Scholar]
- 36.Geddes BA, Pickering BS, Poysti NJ, Collins H, Yudistira H, Oresnik IJ. 2010. A locus necessary for the transport and catabolism of erythritol in Sinorhizobium meliloti. Microbiology 156:2970–2981. doi: 10.1099/mic.0.041905-0. [DOI] [PubMed] [Google Scholar]
- 37.Geddes BA, Oresnik IJ. 2012. Genetic characterization of a complex locus necessary for the transport and catabolism of erythritol, adonitol and L-arabitol in Sinorhizobium meliloti. Microbiology 158:2180–2191. doi: 10.1099/mic.0.057877-0. [DOI] [PubMed] [Google Scholar]
- 38.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krol E, Becker A. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol Genet Genomics 272:1–17. [DOI] [PubMed] [Google Scholar]
- 40.Yuan ZC, Zaheer R, Morton R, Finan TM. 2006. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res 34:2686–2697. doi: 10.1093/nar/gkl365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue Y, Kimura A. 1999. Glycolytic-methylglyoxal pathway. Proc Jpn Acad Ser B 75:127–132. doi: 10.2183/pjab.75.127. [DOI] [Google Scholar]
- 42.Weber J, Kayser A, Rinas U. 2005. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. II. Dynamic response to famine and feast, activation of the methylglyoxal pathway and oscillatory behaviour. Microbiology 151:707–716. [DOI] [PubMed] [Google Scholar]
- 43.Bourdes A. 2008. Role of aminotransferases during the symbiosis between Rhizobium leguminosarum and Pisum sativum. PhD thesis University of Reading, Reading, United Kingdom. [Google Scholar]
- 44.Prell J, Bourdes A, Karunakaran R, Lopez-Gomez M, Poole PS. 2009. Pathway of γ-aminobutyrate metabolism in Rhizobium leguminosarum 3841 and its role in symbiosis. J Bacteriol 191:2177–2186. doi: 10.1128/JB.01714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finnie C, Hartley NM, Findlay KC, Downie JA. 1997. The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol Microbiol 25:135–146. doi: 10.1046/j.1365-2958.1997.4471803.x. [DOI] [PubMed] [Google Scholar]
- 46.Krehenbrink M, Downie JA. 2008. Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae. BMC Genomics 9:55. doi: 10.1186/1471-2164-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cremers HC, Batley M, Redmond JW, Eydems L, Breedveld MW, Zevehuizen LP, Lugtenberg BJ. 1990. Rhizobium leguminosarum exoB mutants are deficient in the synthesis of UDP-glucose 4′-epimerase. J Biol Chem 265:21122–21127. [PubMed] [Google Scholar]
- 48.Janczarek M. 2011. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int J Mol Sci 12:7898–7933. doi: 10.3390/ijms12117898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nentwich SS, Brinkrolf K, Gaigalat L, Hüser AT, Rey DA, Mohrbach T, Kalinowski J. 2009. Characterization of the LacI-type transcriptional repressor RbsR controlling ribose transport in Corynebacterium glutamicum ATCC 13032. Microbiology 155:150–164. doi: 10.1099/mic.0.020388-0. [DOI] [PubMed] [Google Scholar]
- 50.Batista S, Patriarca EJ, Tatè R, Martínez-Drets G, Gill PR. 2009. An alternative succinate (2-oxoglutarate) transport system in Rhizobium tropici is induced in nodules of Phaseolus vulgaris. J Bacteriol 191:5057–5067. doi: 10.1128/JB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glenn AR, Brewin NJ. 1981. Succinate resistant mutants of Rhizobium leguminosarum. Microbiology 126:237–241. doi: 10.1099/00221287-126-1-237. [DOI] [Google Scholar]
- 52.Reid CJ, Poole PS. 1998. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J Bacteriol 180:2660–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronson CW, Astwood PM, Downie JA. 1984. Molecular cloning and genetic organization of C4-dicarboxylate transport genes from Rhizobium leguminosarum. J Bacteriol 160:903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanger A. 2008. Analysis of the ECF sigma factors of Rhizobium leguminosarum biovar viciae 3841. PhD thesis University of East Anglia, Norwich, United Kingdom. [Google Scholar]
- 55.Heimann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110. doi: 10.1016/S0065-2911(02)46002-X. [DOI] [PubMed] [Google Scholar]
- 56.Noel KD, Diebold RJ, Cava JR, Brink BA. 1988. Rhizobial purine and pyrimidine auxotrophs: nutrient supplementation, genetic analysis, and the symbiotic requirement for the novo purine biosynthesis. Arch Microbiol 149:499–506. doi: 10.1007/BF00446751. [DOI] [Google Scholar]
- 57.Lemieux MJ. 2008. A perspective on the structural studies of inner membrane electrochemical potential-driven transporters. Biochim Biophys Acta 1778:1805–1813. doi: 10.1016/j.bbamem.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 58.Hood G, Karunakaran R, Downie JA, Poole PS. 2015. MgtE from Rhizobium leguminosarum is a Mg2+ channel essential for growth at low pH and N2 fixation on specific plants. Mol Plant Microbe Interact 28:1281–1287. doi: 10.1094/MPMI-07-15-0166-R. [DOI] [PubMed] [Google Scholar]
- 59.Sandler SJ, Hugenholtz P, Schleper C, DeLong EF, Pace NR, Clark AJ. 1999. Diversity of radA genes from cultured and uncultured archaea: comparative analysis of putative RadA proteins and their use as a phylogenetic marker. J Bacteriol 181:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos-Beneit F. 2015. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol 6:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 62.Steed PM, Wanner BL. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol 175:6797–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Zhang Y. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob Agents Chemother 51:2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calderón-Flores A, Du Pont G, Huerta-Saquero A, Merchant-Larios H, Servín-González L, Durán S. 2005. The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J Bacteriol 187:5075–5083. doi: 10.1128/JB.187.15.5075-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hecker M, Richter A, Schroeter A, Wölfel L, Mach F. 1987. Synthesis of heat shock proteins following amino acid or oxygen limitation in Bacillus subtilis relA+ and relA strains. Z Naturforsch 42:941–947. [PubMed] [Google Scholar]
- 66.Amar M, Patriarca EJ, Manco G, Bernard P, Riccio A, Lamberti A, Laccarino M. 1994. Regulation of nitrogen metabolism is altered in a glnB mutant strain of Rhizobium leguminosarum. Mol Microbiol 11:685–693. doi: 10.1111/j.1365-2958.1994.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 67.Mulley G, White JP, Karunakaran R, Prell J, Bourdes A, Bunnewell S, Poole PS. 2011. Mutation of GOGAT prevents pea bacteroid formation and N2 fixation by globally downregulating transport of organic nitrogen sources. Mol Microbiol 80:149–167. doi: 10.1111/j.1365-2958.2011.07565.x. [DOI] [PubMed] [Google Scholar]
- 68.Ercolano R, Mirabella R, Chiurazzi M, Merrick M. 2001. The Rhizobium leguminosarum glnB gene is down-regulated during symbiosis. Mol Gen Genet 264:555–564. doi: 10.1007/s004380000333. [DOI] [PubMed] [Google Scholar]
- 69.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tambalo DD, Yost CK, Hynes MF. 2010. Characterization of swarming motility in Rhizobium leguminosarum bv. viciae. FEMS Microbiol Lett 307:165–174. doi: 10.1111/j.1574-6968.2010.01982.x. [DOI] [PubMed] [Google Scholar]
- 71.Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. Microbiology 84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 72.Poole PS, Schofiel NA, Reid CJ, Drew EM, Walshaw DL. 1994. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 73.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 74.Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc 6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 76.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.