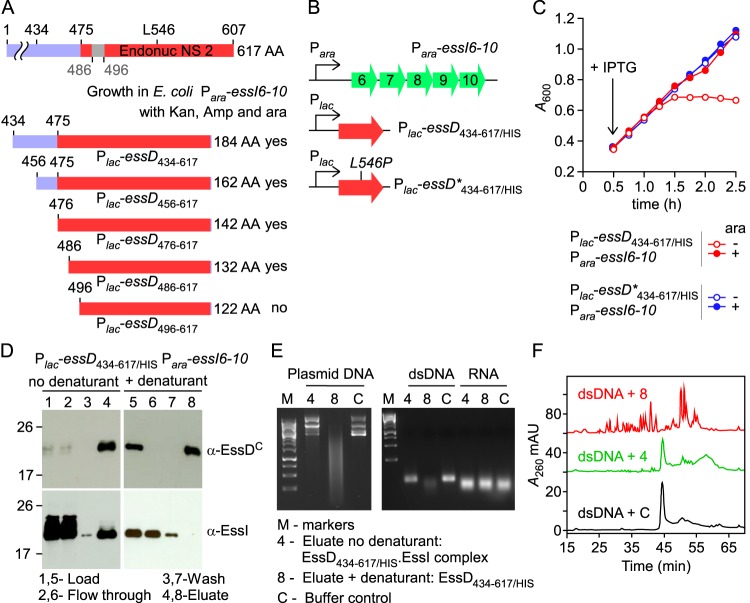
FIG 4.
Biochemical characterization of EssD434–617/HIS. (A) EssD sequence elements required for DNase activity and inhibition by EssI. Plasmids encoding N-terminal truncations of EssD were transformed in E. coli Para-essI6-10 for the selection of viable clones in the presence of kanamycin, ampicillin, and arabinose. (B) Diagram depicting plasmid constructs with lac and ara promoters. (C) Growth of E. coli variants carrying Para-essI6-10 with either Plac-essD434–617/HIS or Plac-essD*434–617/HIS plasmids was monitored as described in the legend to Fig. 2B. (D) Wild-type EssD434–617/HIS was purified from cleared lysate of E. coli Plac-essD434–617/HIS Para-essI6-10. Aliquots of samples prior to purification (Load), flowthrough, wash, and eluates were separated by SDS-PAGE and transferred to PVDF membrane for immunoblotting with anti-EssDC and anti-EssI polyclonal sera. Lanes 1 to 4 and 5 to 8 were loaded with sample aliquots of preparations purified in buffer A without urea (no denaturant) or buffer A containing 8 M urea (+ denaturant), respectively. Bound proteins were eluted in buffer A containing 0.5 M imidazole. Numbers to the left indicate the mobility of molecular mass markers. (E) Plasmid DNA, dsDNA, or RNA was incubated with proteins eluted in fraction 4 or 8 or buffer control. Following incubation, reaction products were separated on an agarose gel and visualized with a UV transilluminator for image acquisition. (F) Reaction products following incubation of dsDNA with sample 4, sample 8, or buffer control C were separated by reverse-phase chromatography, and absorbance was recorded at 260 nm (A260). AU, arbitrary units.