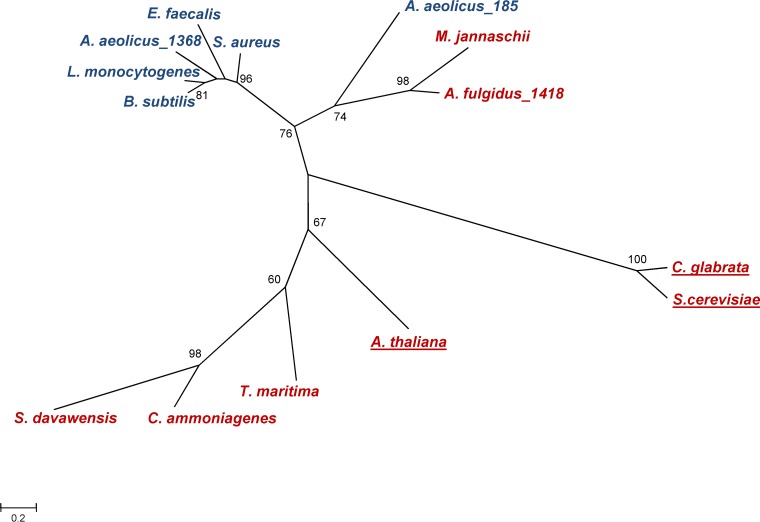
FIG 9.
Phylogenetic tree based on amino acid sequences of glycerol phosphate cytidylyltransferases (GCTs), bifunctional riboflavin kinase/FMN adenylyltransferases (FADS/RibFKs), and flavin adenine dinucleotide synthetases (FADSs), whose functions have been proven. Only the FADS domain of the bifunctional FADS/RibFKs was used for this analysis. MEGA6 software (28) used for sequence alignment and to draw the tree. Bootstrap values were calculated from 1,000 replicates. Only bootstrap values greater than or equal to 60 are shown. The scale bar represents the branch lengths measured in the number of substitutions per site. Organisms harboring the GCT and FADS proteins are indicated in blue and red, respectively. Underlined names refer to eukaryotic organisms. GenPept accession numbers for the FADS and GCT proteins are provided in parentheses for the following organisms: for bifunctional RibFK/FADS, Corynebacterium ammoniagenes (Q59263.1), Thermotoga maritima (1T6Y), and Streptomyces davawensis (ABN55909.1); for FADS, Arabidopsis thaliana (Q9FMW8.1), Methanocaldococcus jannaschii (WP_010870692), and Archaeoglobus fulgidus AF1418 (WP_048064389.1); and for GCT, Aquifex aeolicus AQ1368 (NP_213944.1), Candida glabrata (3G5A), Saccharomyces cerevisiae (AAA65730.1), Bacillus subtilis (1COZ), Aquifex aeolicus AQ185 (NP_213132.1), Staphylococcus aureus (AAB51063.1), Listeria monocytogenes (WP_003727003), and Enterococcus faecalis (EEI58902).