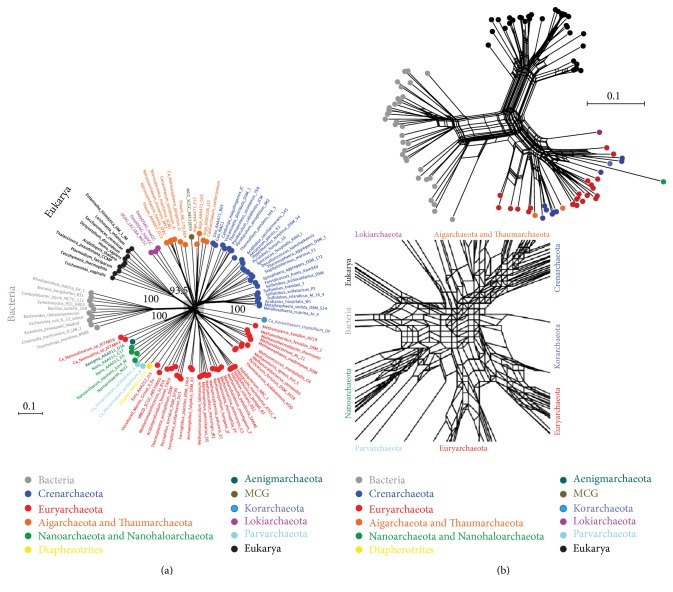
Figure 3.
Distance networks do not support AAS. (a) The concatenated alignment of Spang et al. (2015) was provided by Lionel Guy and Thijs Ettema [1]. This alignment concatenated 36 genes (arCOGs) in 104 taxa (84-10-10 dataset) and was already trimmed by authors to remove sites containing >50% gaps. A splits-tree distance-based network (character sites = 10,547, LS fit = 99.97; δ-score = 0.25) reconstructed from the 84-10-10 dataset does not support AAS [1]. Eukaryotic proteomes are in close proximity to Lokiarchaeota but form a monophyletic group of their own. Numbers on branches indicate BS support values for deep split events. The inset shows reticulations at the base of the tree. MCG: miscellaneous crenarchaeotal group. (b) An unrooted splits-tree distance-based network (character sites = 493, LS fit = 99.61; δ-score = 0.24) reconstructed from 34-34-34 dataset sampled from Archaea (including Lokiarchaeum), Bacteria, and Eukarya and 493 characters corresponding to presence/absence of FSF domains in the universal ABE (459) and AE (34) groups of Figure 1. For this reconstruction, we only considered organisms exhibiting “free-living” lifestyles since parasitic and obligate parasitic organisms tend to have reduced genomes that are distorted by their holobiont relationship biasing the data matrix. The only “non-free-living” exception was Nanoarchaeota that was added to ensure consistency with the 84-10-10 dataset and to maximize the coverage of archaeal phyla [1]. Both unrooted networks reconstructed by SplitsTree (ver. 4.13.1) [50].