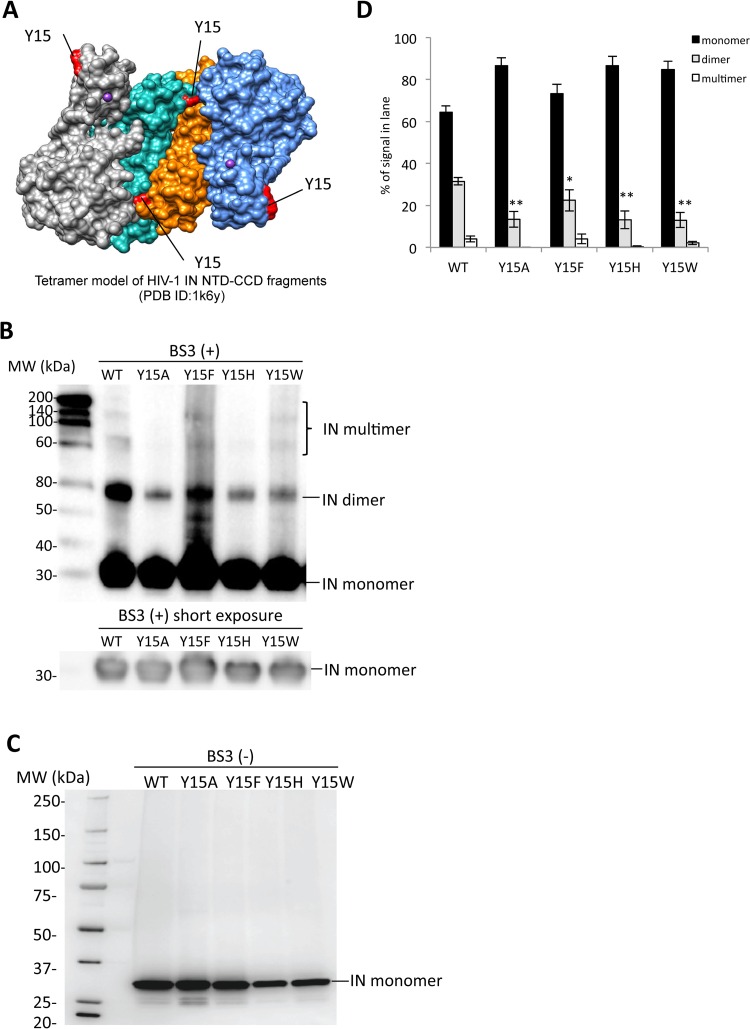
FIG 5.
Biochemical properties of the Tyr15 mutant IN. (A) The tetramer structure for the HIV-1 IN NTD-CCD fragment PDB ID (1k6y) (31) was created by using the UCSF Chimera program. The locations of the Tyr15 residues are indicated in red. (B) Twenty nanograms of each recombinant IN protein was cross-linked by BS3, and the resulting products were analyzed by Western blotting with mouse monoclonal anti-IN antibody (ab66645; Abcam). The gel fraction corresponding to the IN monomer with shorter exposure is shown below. (C) A representative blot for each rIN without BS3 is shown as a control. (D) Signal intensities of IN monomer, dimer, and multimer were quantified by using ImageJ software. The relative intensity (as a percentage) of each signal is shown; total signal intensity of each lane was set as 100%. Bars represent the mean results ± standard deviations. The asterisk indicates the P value for each mutant versus WT by two-tailed Student's t test (*, P < 0.05; **, P < 0.01).