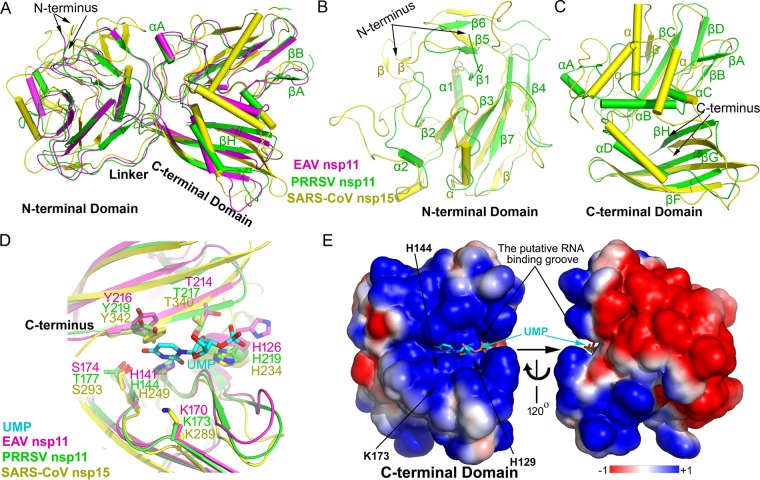
FIG 2.
Structural superimposition, catalytic site, and electrostatic potential of PRRSV nsp11, EAV nsp11, and SARS-CoV nsp15. Shown is a structural alignment of the whole protein (A), NTD (B), and CTD (C). EAV nsp11, magenta; PRRSV nsp11, green; SARS-CoV nsp15, yellow. (D) Active-site grooves in the CTD of NendoU from different nidoviruses. (E) Electrostatic potential of PRRSV nsp11 and a 120°-rotated view (right). The surface potential is displayed as a color gradient ranging from red (negative) to blue (positive). Note that three putative catalytic residues involving Lys173, His144, and His129 are located in a groove formed between β-sheet βH and a four-stranded β-sheet (βA, βB, βD, and βC). The modeled uridine 3′-phosphate (UMP) molecule is shown in a ball-and-stick representation. Note that both complete protomers of PRRSV nsp11 and EAV nsp11 are used.