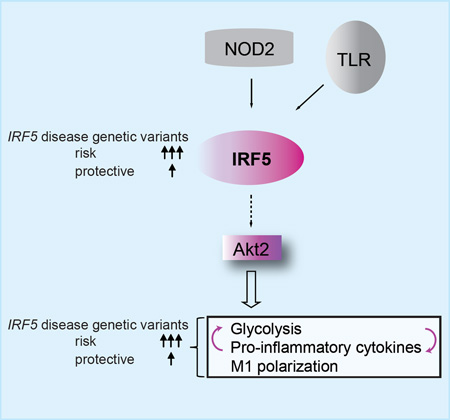
Summary
Interferon regulatory factor 5 (IRF5) regulates inflammatory M1 macrophage polarization, and disease-associated IRF5 genetic variants regulate pattern-recognition-receptor (PRR)-induced cytokines. PRR-stimulated macrophages and M1 macrophages exhibit enhanced glycolysis, a central mediator of inflammation. We find that IRF5 is needed for PRR-enhanced glycolysis in human macrophages and in mice in vivo. Upon stimulation of the PRR nucleotide binding oligomerization domain containing 2 (NOD2) in human macrophages, IRF5 binds RIP2, IRAK1 and TRAF6. IRF5, in turn, is required for optimal Akt2 activation, which increases expression of glycolytic pathway genes and HIF1A, as well as pro-inflammatory cytokines and M1 polarization. Furthermore, pro-inflammatory cytokines and glycolytic pathways co-regulate each other. Rs2004640/rs2280714 TT/TT IRF5 disease-risk-carrier cells demonstrate increased IRF5 expression and increased PRR-induced Akt2 activation, glycolysis, pro-inflammatory cytokines and M1 polarization relative to GG/CC carrier macrophages. Our findings identify that IRF5 disease-associated polymorphisms regulate diverse immunological and metabolic outcomes and provide further insight into mechanisms contributing to the increasingly recognized important role for glycolysis in inflammation.
Graphical Abstract
eTOC Blurb
Hedl et al. find that IRF5 is a critical link to pattern-recognition receptor (PRR)- and M1 human macrophage-enhanced glycolysis; this link requires Akt2 activation. IRF5 also regulates PRR-induced glycolysis in vivo in mice. IRF5 disease-risk variants show increased Akt2 activation, glycolysis, M1 polarization and pro-inflammatory cytokines.
Introduction
Proper pattern recognition receptor (PRR) responses to microbial exposure are crucial in maintaining mucosal immune homeostasis. Most studies show that IRF5 is critical for optimal cytokine secretion upon stimulation of a broad range of PRRs (Hedl and Abraham, 2012; Krausgruber et al., 2011; Takaoka et al., 2005), although other studies show more PRR selectivity (Bergstrom et al., 2015). IRF5 polymorphisms resulting in increased IRF5 transcripts (Graham et al., 2006) are associated with multiple immune-mediated diseases, including systemic lupus erythematosus (SLE), inflammatory bowel disease (IBD), rheumatoid arthritis, Sjogren's syndrome, primary biliary cirrhosis, systemic sclerosis and multiple sclerosis (Lazzari and Jefferies, 2014). Moreover, we found that IRF5 disease-associated polymorphisms dramatically affect inter-individual variance across the population in PRR-induced cytokines in myeloid cells (Hedl and Abraham, 2012), thereby raising the possibility that these polymorphisms may contribute to differences in additional PRR-initiated outcomes. As a transcription factor IRF5 binds to cytokine gene promoters to enhance PRR-initiated cytokine expression (Barnes et al., 2002; Krausgruber et al., 2011; Saliba et al., 2014; Takaoka et al., 2005). IRF5 also directly binds the adaptor molecule MyD88 and the signaling intermediates IRAK1 and TRAF6 in overexpression systems and mouse macrophages (Balkhi et al., 2008; Inoue et al., 2014; Takaoka et al., 2005). How IRF5 regulates downstream pathways is incompletely understood. We found that IRF5 is required for optimal PRR-initiated mitogen-activated protein kinase (MAPK) and NF-kB activation in human myeloid cells (Hedl and Abraham, 2012), although IRF5 did not regulate these pathways in mouse B cells (Takaoka et al., 2005). This could be due to differences in cell types, but also to differences in human and mouse inflammatory responses, which can be quite distinct (Seok et al., 2013), highlighting the importance of defining IRF5 regulation in primary human cells. Consistent with its role in pathogen responses (Pandey et al., 2009), most (Dalmas et al., 2015; Krausgruber et al., 2011), but not all (Lacey et al., 2012) studies find that IRF5 is required for the pro-inflammatory M1 macrophage phenotype.
Interestingly, M1 macrophages show increased glycolytic flux (Ganeshan and Chawla, 2014). Whether IRF5 is required for glycolysis has not been examined. Glycolysis is a central mediator of inflammatory outcomes, cell maturation, cytokine secretion and bacterial clearance in myeloid cells (Cramer et al., 2003; Everts et al., 2012; Krawczyk et al., 2010; O'Neill and Hardie, 2013), and regulates immune responses in vitro and in vivo in additional cell types (O'Neill and Hardie, 2013). Following PRR stimulation, glycolysis maintains mitochondrial ATP production and cell viability (Cramer et al., 2003; Everts et al., 2012). Despite the importance of IRF5 variants in human inflammatory diseases and of IRF5 in cytokine secretion in PRR-stimulated myeloid cells, a number of questions remain unanswered. What are the mechanisms through which IRF5 modulates human macrophage polarization? Given that IRF5 regulates PRR-initiated cytokines and M1 polarization, and the critical role of enhanced glycolysis under both these situations, does IRF5 regulate glycolysis, and if so, through what mechanisms? As the risk and non-risk alleles of these variants are commonly distributed among the population, is M1 polarization and/or glycolysis regulated in an IRF5 genotype-dependent manner?
In this study, in primary human monocyte-derived macrophages (MDMs), we found that IRF5 was essential for PRR-initiated upregulation of glycolysis. We uncover that with PRR stimulation, IRF5 leads to Akt2 activation, which results in enhanced glycolysis and M1 polarization, and we dissect distinct upstream and downstream regulation of IRF5-dependent signaling pathways. We further determine that IRF5-disease risk variants commonly expressed in the population regulate increased inflammation through increased Akt2 activation, glycolysis, and M1 polarization, thereby highlighting mechanisms by which IRF5 regulates immune-mediated diseases.
Results
PRR stimulation enhances glycolysis through IRF5 signaling
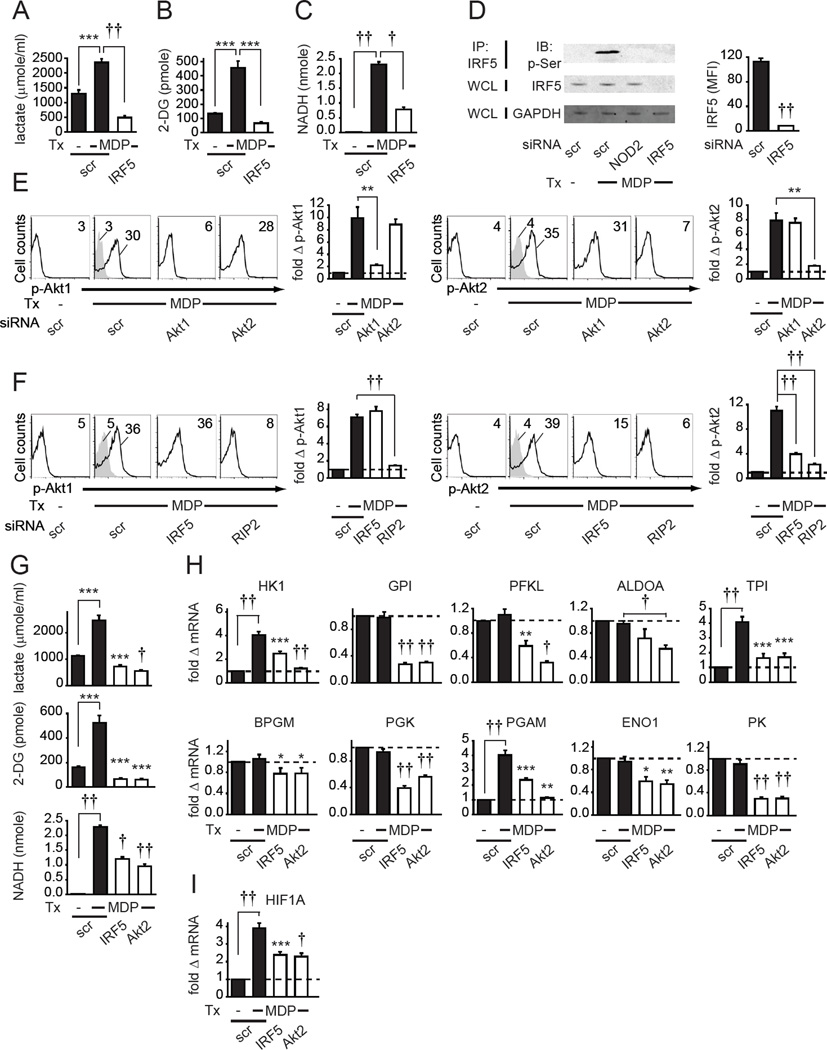
TLR stimulation induces glycolysis in human and mouse myeloid cells (Cramer et al., 2003; Marsin et al., 2002; Rodriguez-Prados et al., 2010), and glycolysis is essential for various immune outcomes in vitro and in vivo (Cramer et al., 2003; Krawczyk et al., 2010; O'Neill and Hardie, 2013). Given that IRF5 regulates PRR-initiated cytokine secretion in human myeloid-derived cells, we hypothesized that IRF5 would be necessary for PRR-induced glycolysis in human macrophages. We first confirmed that PRR stimulation increases glycolysis. We stimulated MDMs with muramyl dipeptide (MDP), the minimal bacterial peptidoglycan component that specifically activates NOD2, a PRR associated with inflammatory bowel disease (IBD) (Abraham and Cho, 2009), and found that lactate secretion, a measure of glycolytic flux, was increased (Fig. 1A). Glucose uptake, which is also associated with glycolysis, was similarly increased (Fig. 1B). These outcomes were associated with a direct increase in glycolytic pathway activity; NOD2 stimulation increased hexokinase activity, the first step, and a rate limiting glycolytic enzyme (Fig. 1C). We next effectively knocked down IRF5 (Fig 1D and Fig. S1A), and found that IRF5 deficiency led to decreased lactate production, glucose uptake and hexokinase activity in NOD2-stimulated MDMs (Fig. 1A–C). Furthermore, following PRR stimulation of myeloid cell lines, IRF5 can be phosphorylated and this modification is required for certain downstream outcomes (Lopez-Pelaez et al., 2014; Ren et al., 2014). MDP treatment led to IRF5 phosphorylation in a NOD2-dependent manner as measured by a phospho-serine motif detecting antibody (Fig. 1D). Taken together, IRF5 is required for NOD2-induced glycolysis.
Fig. 1. NOD2-induced IRF5 and Akt2 signaling regulates glycolytic pathway genes and glycolysis in human MDMs.
(A–C) Human MDMs (n=4 donors) were transfected with IRF5 siRNA, then treated for 24h with 100µg/ml MDP. (A) Lactate production. (B) Glucose uptake. (C) Hexokinase activity. Mean+SEM. (D) MDMs were transfected with the indicated siRNA. (Left) Following 15min 100µg/ml MDP treatment, IRF5 was immunoprecipitated from cell lysates and IRF5 serine phosphorylation was assessed by western blot. IRF5 and GAPDH expression from total lysates served as loading controls. (Right) Summary graph with mean fluorescence intensity (MFI) values for IRF5 expression assessed by flow cytometry+SEM (n=4). (E–F) MDMs (n=8 for each panel) were transfected with the indicated siRNA and then treated for 15min with 100µg/ml MDP. Representative flow cytometry with MFI values for phospho-kinase, and summary graph with fold phospho-kinase, induction compared to untreated, scrambled siRNA-transfected cells for each donor+SEM. Isotype controls (gray shading). (G) MDMs (n=4) were transfected with IRF5 or Akt2 siRNA and then treated with 100µg/ml MDP for 24h. Lactate production, glucose uptake and hexokinase activity. Mean+SEM. (H–I) MDMs (n=8) were transfected with IRF5 or Akt2 siRNA and then treated with 100µg/ml MDP for 4h. Fold mRNA expression compared to untreated, scrambled siRNA-transfected cells for each donor+SEM. Similar results were seen in an additional n=4 for (A–C, E–F, G) and n=8 for (H–I). (G–I) Significance relative to scrambled siRNA-transfected, MDP-treated cells or as indicated. Tx, treatment; scr, scrambled. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
IRF5 is required for Akt2 activation and Akt2 mediates NOD2-induced glycolysis
Akt2 signaling regulates glycolysis in mouse hepatocytes (Panasyuk et al., 2012). To our knowledge, Akt2 contributions to glycolysis have not been reported in human MDMs. We therefore asked if IRF5 regulates NOD2-induced Akt2 activation using a phospho-Akt2 specific antibody. As a control, we measured Akt1 phosphorylation. In mouse cells Akt1 and Akt2 contribute to M2 and M1 polarization, respectively (Arranz et al., 2012), and we similarly found that Akt1 was required for optimal M2 polarization in human MDMs (data not shown). M1 macrophages show increased glycolysis, and M2 macrophages show decreased glycolysis relative to non-polarized macrophages (Rodriguez-Prados et al., 2010). We first established that NOD2 stimulation induced both Akt1 and Akt2 phosphorylation in MDMs (Fig. 1E). We ensured that phospho-Akt1 and phospho-Akt2 antibodies detected their specific targets using a siRNA approach (Fig. 1E). We confirmed reduced phospho-Akt1 and phospho-Akt2 with knockdown through an independent approach (Fig. S1B). Interestingly, IRF5 knockdown decreased NOD2-induced Akt2 activation, but not Akt1 activation (Fig. 1F). Neither NOD2 stimulation (Fig. S1C) nor IRF5 knockdown affected total Akt2 levels (Fig. S1D). However, RIP2, an adapter molecule required for NOD2 signaling (Abraham and Cho, 2009), was required for both NOD2-induced Akt1 and Akt2 activation (Fig. S1E & Fig. 1F). Furthermore, Akt2 knockdown decreased NOD2-induced glycolytic flux to similar levels as that observed for IRF5 knockdown (Fig. 1G). We confirmed these results with additional independent siRNAs targeting IRF5 and Akt2 (Fig. S1F&G). Moreover, IRF5 and Akt2 regulated glycolysis downstream of additional PRRs in MDMs, including TLR2, TLR3 and TLR4 (Fig. S1H). We confirmed that each PRR ligand specifically stimulated its respective receptor (Fig. S1I); glycolysis remained intact in these same MDMs stimulated by a distinct PRR ligand (data not shown). Furthermore, IRF5 knockdown did not affect expression of the examined PRRs and PRR knockdown did not alter IRF5 expression (Fig. S1J–K).
We next sought to define mechanisms through which IRF5 regulates NOD2-induced glycolysis. We examined genes in the glycolysis pathway that convert glucose to pyruvate (Munoz-Pinedo et al., 2012) and found that NOD2 stimulation increased hexokinase (HK1), triosephosphate isomerase (TPI) and phosphoglycerate mutase (PGAM), but not glucose-6-phosphate isomerase (GPI), phosphofructokinase (PFKL), aldolase (ALDOA), bisphosphoglycerate mutase (BPGM), phosphoglycerate kinase (PGK), enolase (ENO1) and pyruvate kinase (PK) expression (Fig. 1H). However, optimal expression of both induced and non-induced genes mostly required IRF5 in NOD2-stimulated MDMs (Fig. 1H). Akt2 was similarly required for optimal expression of the glycolytic genes assessed (Fig. 1H). IRF5 and Akt2 further contributed to NOD2-induced HIF1A expression (Fig. 1I), which also regulates glycolysis in macrophages (Cramer et al., 2003). Taken together, IRF5 mediates Akt2 activation, and both molecules are required for the optimal basal and NOD2-induced expression of multiple glycolytic pathway genes and HIF1A.
IRF5 regulates glycolysis in mice in vivo
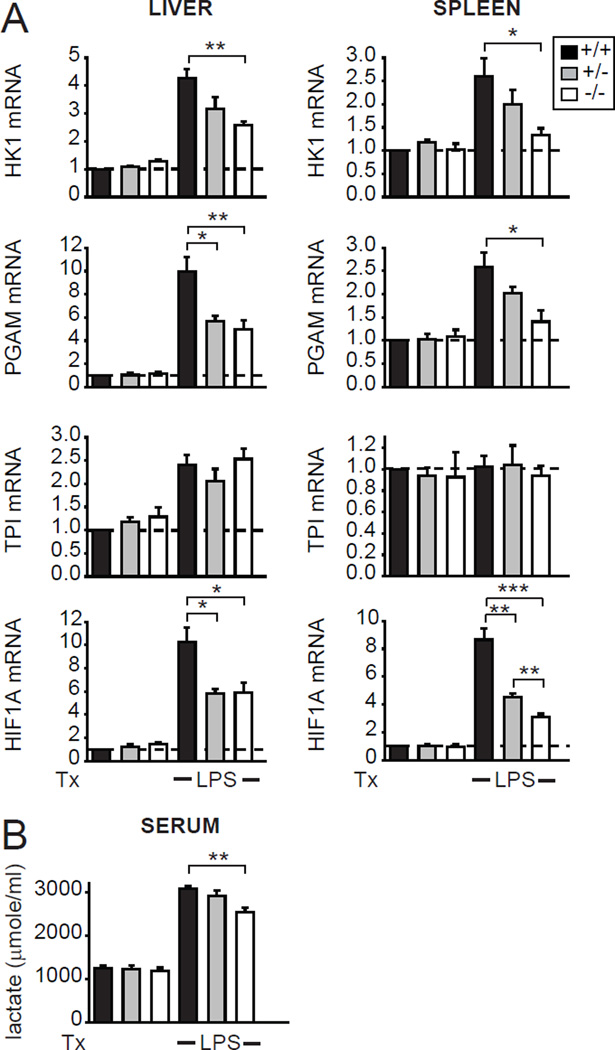
We next questioned if IRF5 regulates PRR-induced glycolysis in vivo. Upon LPS injection, the induction of HK1, PGAM and HIF1A in liver and spleen (Fig. 2A) and of serum lactate (Fig. 2B) were significantly decreased in IRF5-deficient mice relative to WT controls. Therefore, IRF5 regulates glycolysis in mice in vivo.
Fig. 2. IRF5-deficient mice exhibit decreased LPS-induced glycolysis in vivo.
IRF5+/+, IRF5+/− or IRF5−/− mice (n=4–5/genotype) were injected i.p. with 10µg LPS for 24h. (A) Fold mRNA expression in liver and spleen, and (B) serum lactate levels (representative of 2 independent experiments). Tx, treatment; *, p<0.05; **, p<0.01; ***, p<0.001.
Glycolytic pathway genes regulate NOD2-induced glycolysis and HIF1A expression
We next asked if the IRF5-dependent glycolytic-pathway genes we had identified were, in fact, necessary for NOD2-induced glycolysis in MDMs, and questioned if these glycolytic pathway genes co-regulated each other. We selected the subset of genes whose expression was induced with NOD2 stimulation, including HK1, TPI and PGAM (Fig. 1H) and HIF1A (Fig. 1I), effectively knocked down their transcript and protein expression (Fig. S2A&B), and ensured the cells were viable (Fig. S2C). We also ensured that HK1 knockdown diminished hexokinase activity (data not shown). Each of these genes was required for NOD2-induced glycolysis (Fig. S2D). Importantly, knockdown of each of the glycolytic genes decreased NOD2-induced expression of the other glycolytic genes and of HIF1A and vice versa (Fig. S2A); the effect was more pronounced following combined silencing of the three glycolytic genes (Fig. S2A). IRF5 and Akt2 expression was not affected by knockdown of these glycolytic genes (Fig. S2E). However, knockdown of each of the glycolytic genes decreased Akt2 activation to varying degrees, while Akt1 activation remained unchanged (Fig. S2E), thereby demonstrating selectivity in the regulation of downstream pathway activation and that Akt2 activation and glycolytic genes can co-regulate each other. Consistently, the regulation of Akt activation by glycolysis has been observed in cancer cells and overexpression systems (Pelicano et al., 2006). HIF1A expression is regulated both following PRR stimulation and under hypoxic conditions (Sica and Mantovani, 2012). We found that inducing hypoxia through two independent approaches also induced Akt2 activation, glycolytic gene expression and glycolysis in an IRF5-dependent manner in human MDMs (Fig. S2F–H); we ensured that the cells were viable under these conditions (Fig. S2I). Taken together, NOD2-induced glycolytic pathway genes and HIF1A are required for optimal NOD2-induced glycolysis in MDMs, and these genes regulate the expression of each other.
IRF5 regulates the increased glycolysis observed in M1 macrophages, and Akt2 and MAPK/NF κB signaling show distinct regulation of pro- and anti-inflammatory cytokines
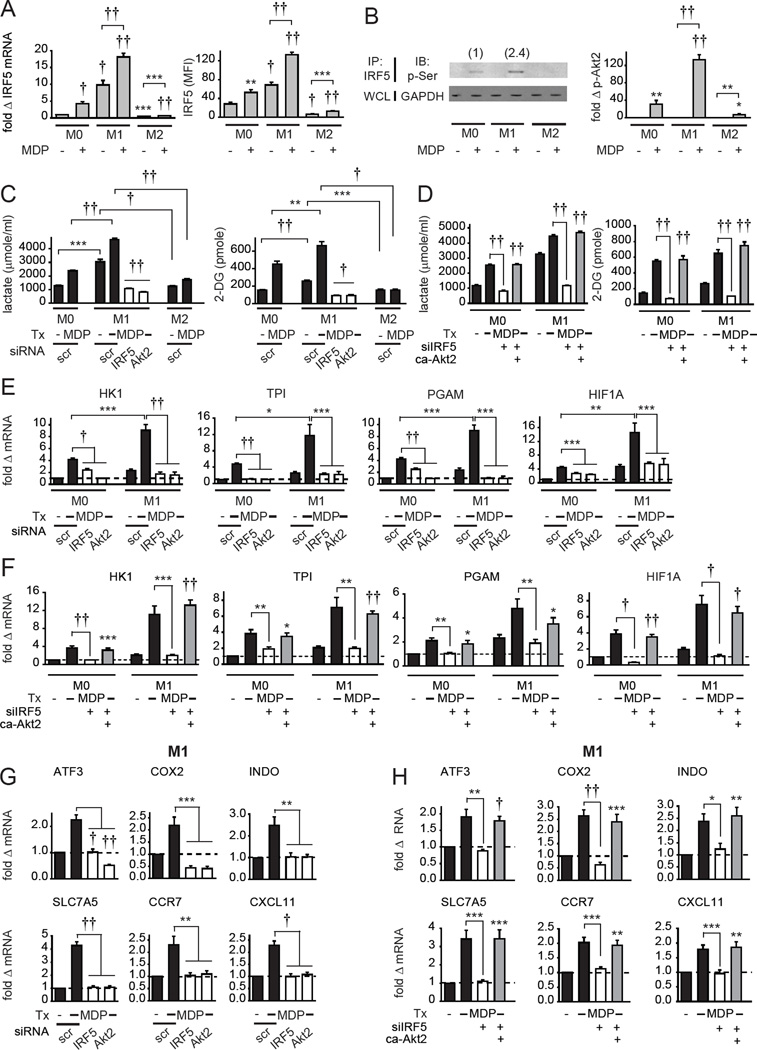
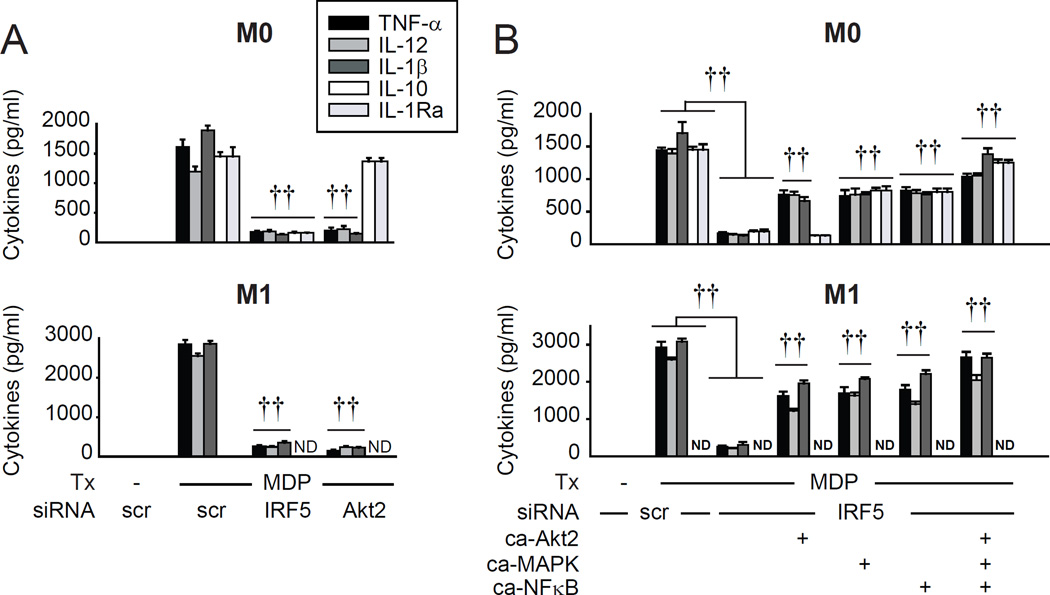
Given that M1 macrophages exhibit increased glycolysis (Ganeshan and Chawla, 2014) and increased glycolytic gene expression (Galvan-Pena and O'Neill, 2014), and that IRF5 contributes to M1 polarization (Krausgruber et al., 2011), we next examined how IRF5 regulates glycolysis in distinct macrophage subtypes. While macrophage polarization involves a broad spectrum of phenotypes, we differentiated MDMs into M1 and M2 phenotypes utilizing common conditions, and confirmed that they expressed appropriate markers as previously described for human cells (Martinez et al., 2006) compared to the non-polarized (M0) MDMs used in the studies above (Fig. S3A). We confirmed that both IRF5 mRNA and protein were upregulated in M1 macrophages relative to M0 macrophages (Fig. 3A). Further, IRF5 expression and phosphorylation, and Akt2 activation were increased following NOD2 stimulation (Fig 3A&B), with activation higher in M1 macrophages than that in undifferentiated macrophages (Fig. 3B). In contrast, these outcomes were dramatically reduced in M2 macrophages (Fig. 3A&B). Compared to non-polarized macrophages, M1 macrophages showed increased glycolysis that was further increased with NOD2 stimulation (Fig. 3C). The TLR4 ligand lipid A enhanced glycolysis through IRF5 (Fig. S1H). Consistently, the TLR4 ligand LPS used for M1 differentiation activated Akt2 and enhanced glycolysis in an IRF5-dependent manner (Fig. S3B&C). Therefore, impaired responses to LPS during M1 skewing conditions are likely one mechanism contributing to the M1 polarization defect in IRF5-deficient MDMs. Similar to non-polarized MDMs (Fig. 1), optimal glycolysis in M1 macrophages required IRF5 and Akt2 (Fig. 3C). Compared to M1 macrophages, glycolysis was reduced in M2 macrophages (Fig. 3C). Expressing IRF5 in M0 and M2 macrophages to the levels observed in M1 macrophages (Fig. S3D) increased glycolysis to M1 levels (Fig. S3E). Given the low IRF5 expression and phosphorylation, and low levels of glycolysis in M2 macrophages (Fig. 3A–C), we focused our subsequent studies on M1 macrophages, and occasionally show non-polarized M0 macrophages for comparison in select measures. As Akt2 was necessary for NOD2-induced glycolysis (Fig. 1G and Fig. 3C) and Akt2 activation was IRF5-dependent (Fig. 1F), we restored Akt2 activation to physiological levels in IRF5-deficient MDMs through transfection of constitutively active Akt2 (Fig. S3F). This was sufficient to restore diminished glycolysis in NOD2-stimulated IRF5-deficient non-polarized and M1 macrophages (Fig. 3D). Consistent with the increased glycolysis in M1 macrophages (Fig. 3C), glycolytic pathway genes were increased in M1 relative to M0 macrophages (Fig. 3E). Similar to M0 macrophages, optimal NOD2-induced glycolytic gene expression in M1 macrophages required both IRF5 and Akt2 (Fig. 3E). Consistent with the rescue of glycolysis, restoring Akt2 activation rescued NOD2-induced HK1, PGAM, TPI and HIF1A expression in IRF5-deficient M0 and M1 macrophages (Fig. 3F). We verified the IRF5 contribution to M1 polarization by assessing M1 marker expression (Fig. 3G) and NOD2-induced pro-inflammatory cytokines (Fig. 4A) under IRF5 knockdown conditions. IRF5 was required for both pro- and anti-inflammatory cytokine secretion in non-polarized human myeloid cells upon NOD2 stimulation (Hedl and Abraham, 2012), which we also confirmed here (Fig. 4A); IRF5 was similarly required for optimal IFNα secretion (data not shown). Akt2 signaling was also required for M1 polarization (Fig. 3G) and NOD2-induced pro-inflammatory cytokines (Fig. 4A). Restoring Akt2 activation rescued M1 macrophage markers in IRF5-deficient M1 macrophages (Fig. 3H) and partially rescued pro-inflammatory cytokines in M0 and M1 macrophages (Fig. 4B), indicating that additional pathways contribute to pro-inflammatory cytokines. However, Akt2 was neither necessary nor sufficient for NOD2-induced anti-inflammatory cytokine secretion in M0 macrophages (Fig. 4A&B), indicating that IRF5-dependent signaling pathways other than Akt2 are required for NOD2-induced anti-inflammatory cytokines. Of note is that anti-inflammatory cytokines are already dramatically reduced in M1 relative to non-polarized macrophages, such that anti-inflammatory cytokines were not differentially regulated in the absence of IRF5 or Akt2 in M1 macrophages (Fig 4A).
Fig. 3. NOD2-induced glycolysis in M1 macrophages and M1 polarization is IRF5- and Akt2-dependent.
(A–B) MDMs were left non-polarized (M0) or were polarized to M1 or M2 macrophages as in Experimental Procedures and then treated with 100µg/ml MDP. (A) (Left) IRF5 mRNA (at 4h) (n=4 donors) and (right) protein expression by flow cytometry (at 24h) (n=8). Mean+SEM. (B) (Left) IRF5 phosphorylation at 15min (numbers above bands represent relative band density normalized to treated M0 macrophages), and (right) fold phospho-Akt2 induction by flow cytometry at 15min compared to untreated cells+SEM (n=8). Significance relative to untreated M0 macrophages or as indicated. (C–H) MDMs were transfected with IRF5 or Akt2 siRNA ± empty vector (EV) or constitutively active Akt2 (ca-Akt2), left nonpolarized (M0) or polarized to M1 or M2 for 24h. (C,D) Cells (n=4) were left untreated or treated with 100µg/ml MDP for 24h. (Left) lactate production and (right) glucose uptake. Mean+SEM. (C) For M1 cells significance is compared to scrambled siRNA-transfected, MDP-treated M1 cells or as indicated. (E,F) Cells (n=8) were treated with 100µg/ml MDP for 4h. Fold mRNA induction normalized to untreated, non-polarized, scrambled siRNA-transfected cells for each donor+SEM. (G,H) Fold mRNA induction of M1 markers normalized to untreated, scrambled siRNA-transfected M1 macrophages for each donor+SEM (n=8). Significance relative to empty vector, IRF5 siRNA-transfected cells treated with MDP or as indicated. Tx, treatment; scr, scrambled. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
Fig. 4. NOD2-induced Akt2 signaling regulates pro-inflammatory cytokines, whereas MAPK and NFκB signaling regulates both pro- and anti-inflammatory cytokines.
MDMs or M1 macrophages were transfected with (A) IRF5 or Akt2 siRNA (n=4 donors), or (B) IRF5 siRNA ± empty vector (EV), constitutively active Akt2 (ca-Akt2), a combination of constitutively active ERK, p38 and JNK kinases (ca-MAPK), or constitutively active NFκB (ca-NFκB) (n=8) and treated with 100µg/ml MDP for 24h. Cytokine secretion+SEM. Similar results were observed in an additional n=8. (B) Significance relative to empty vector, IRF5 siRNA-transfected cells treated with MDP or as indicated. Tx, treatment; scr, scrambled; ND, not detected. ††, p<1×10−5.
We previously showed that IRF5 activates PRR-induced MAPK and NFκB pathways (Hedl and Abraham, 2012). We therefore questioned if the MAPK and/or NFκB pathways might mediate NOD2-induced anti-inflammatory cytokine secretion downstream of IRF5 given that IRF5-mediated Akt2 activation did not. We further questioned if MAPK and/or NFκB pathways cooperate with Akt2 to more optimally induce pro-inflammatory cytokines upon NOD2 stimulation. Restoring MAPK and NFκB activation in M0 macrophages through expressing constructs resulting in constitutive activation of these pathways (Fig. S3G&H) partially rescued both pro-inflammatory and anti-inflammatory cytokines (Fig. 4B); rescue of NOD2-induced cytokines was greater with restoring MAPK, NFκB and Akt2 activation in combination (Fig. 4B). Restoring all three pathways in combination in M1 macrophages also more completely rescued the high levels of NOD2-induced pro-inflammatory cytokines (Fig. 4B). Taken together, upon NOD2 stimulation, the different signaling pathways downstream of IRF5 regulate distinct outcomes; Akt2 activation is necessary for glycolysis, M1 polarization and pro-inflammatory cytokine secretion, and the MAPK and NFκB pathways contribute to both pro- and anti-inflammatory cytokine secretion.
The MAPK and NFκ B pathways, and the Akt2 pathway downstream of NOD2 are regulated by IRAK1/TRAF6 and BCAP signaling, respectively
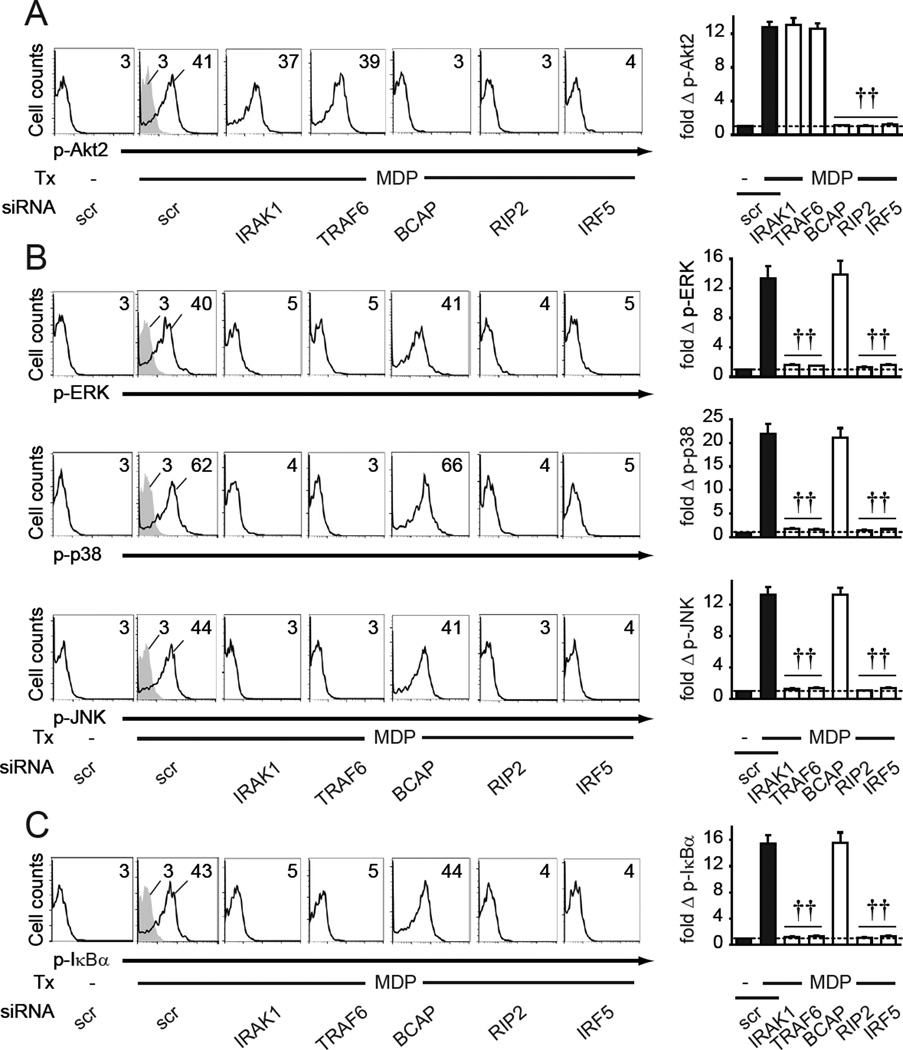
Given the distinct immune outcomes we identified downstream of the NOD2-induced, IRF5-dependent signaling pathways, we next sought to more clearly define the upstream NOD2 signaling intermediates that regulate the activation of the Akt2, MAPK, and NFκB pathways. We (Hedl et al., 2007) and others (Opitz et al., 2004) have found that NOD2 activates IRAK1. IRAK1 activates MAPK and NFκB pathways downstream of PRRs (Kawai and Akira, 2011; Opitz et al., 2004); it is unknown if IRAK1 regulates NOD2-induced Akt2 signaling. We therefore successfully knocked down IRAK1 and TRAF6 (Fig. S4A&B), with which IRAK1 forms a signaling complex (Kawai and Akira, 2011). Signaling by IRAK1 and TRAF6 was not required for NOD2-induced Akt2 activation in MDMs (Fig. 5A). In contrast, IRAK1 and TRAF6 were required for NOD2-induced MAPK (Fig. 5B) and NFκB (Fig. 5C) activation. BCAP is required for PI3K activation through MyD88 in TLR-stimulated myeloid cells (Ni et al., 2012; Troutman et al., 2012). It is unknown if BCAP contributes to NOD2 signaling, where signaling occurs through the distinct adaptor molecule RIP2. We knocked down BCAP (Fig. S4A&B), and found that in contrast to IRAK1 and TRAF6, BCAP was not required for NOD2-induced MAPK and NFκB pathway activation (Fig. 5B&C), but was required for Akt2 activation (Fig. 5A). RIP2 and IRF5 were required for NOD2-initiated signaling through all three pathways (Fig. 5). Therefore, upon NOD2 stimulation, activation of the IRF5-dependent MAPK and NFκB pathways and Akt2 pathway are regulated by distinct branch points, with IRAK1/TRAF6 upstream of MAPK and NFκB activation and BCAP upstream of Akt2 activation.
Fig. 5. IRAK1/TRAF6 and BCAP signaling show distinct regulation of NOD2-induced MAPK and NFκB pathways, and Akt2 pathways, respectively.
MDMs (n=8 donors) were transfected with the indicated siRNA. Cells were then treated for 15min with 100 µg/ml MDP and assessed for (A) Akt2, (B) MAPK, or (C) NFκB pathway activation. (Left) Representative flow with MFI values for phospho-kinases. (Right) Fold phospho-kinase induction compared to untreated, scrambled siRNA-transfected cells for each donor+SEM. Significance relative to MDP-treated, scrambled siRNA-transfected cells. Isotype controls (gray shading). Tx, treatment; scr, scrambled. ††, p<1×10−5.
The transcription factors SP1 and PU.1 bind to glycolytic gene promoters
IRF5 is both a transcription factor and regulator of proximal signaling. We therefore sought to dissect whether IRF5 could regulate PRR-induced glycolytic outcomes by directly binding to the promoters of the glycolytic genes and HIF1A, but found this was not the case (Fig. S4C); such binding has similarly not been reported in studies examining the IRF5 cistrome (Saliba et al., 2014; Wang et al., 2013). NOD2 can also activate IRF3, another transcription factor that can lead to pro-inflammatory outcomes (Sabbah et al., 2009). IRF3 was activated following NOD2 stimulation; however, IRF5 deficiency did not affect IRF3 activation (Fig. S4D). Furthermore, unlike IRF5, IRF3 knockdown did not alter Akt2 activation or glycolysis following NOD2 stimulation (Fig. S4E&F).
We then hypothesized that Akt2-activated transcription factors might be regulating glycolytic gene expression; SP1 and PU.1 can be activated downstream of Akt signaling (Pore et al., 2004; Rieske and Pongubala, 2001) and we identified putative binding sites for these transcription factors in the glycolytic gene promoters based on JASPAR. SP1 and PU.1 binding to the promoters of the glycolytic genes and HIF1A increased upon NOD2 stimulation (Fig. S4G). Furthermore, SP1 and PU.1 knockdown (Fig. S4H) demonstrated that these transcription factors were required for optimal NOD2-induced expression of glycolytic genes (Fig. S4I) and glycolysis (Fig. S4J). Taken together, rather than directly acting in its capacity as a transcription factor, IRF5 likely regulates glycolysis through modulation of Akt2 signaling, which leads to increased binding of Akt-dependent transcription factors to glycolytic gene promoters.
IKKβ, RIP2, IRAK1 and TRAF6 are required for IRF5 phosphorylation and RIP2, IRAK1 and TRAF6 associate with IRF5 following NOD2 stimulation
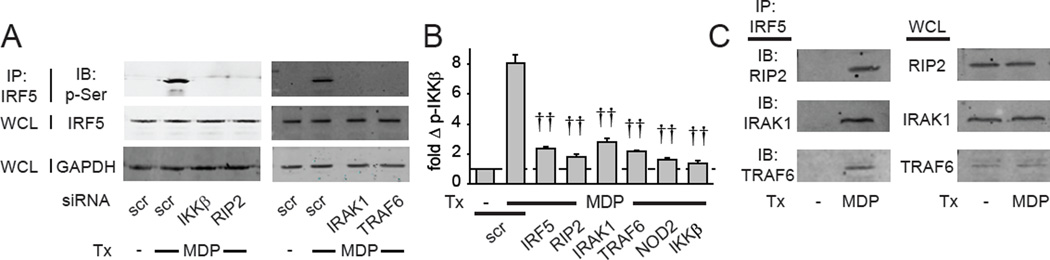
We next sought to better define both the upstream mechanisms regulating IRF5 phosphorylation and the mechanisms through which IRF5 regulates proximal signaling. IKKβ regulates IRF5 phosphorylation in myeloid cell lines (Lopez-Pelaez et al., 2014; Ren et al., 2014), and we found that both IKKβ and the NOD2-adapter molecule RIP2 were required for NOD2-mediated IRF5 serine phosphorylation in human MDMs (Fig. 6A). Following phosphorylation, IRF5 can translocate into the nucleus (Chang Foreman et al., 2012), and we observed peak IRF5 nuclear translocation 4h after NOD2 stimulation (Fig. S5A). Consistent with regulating IRF5 phosphorylation, IKKβ and RIP2 were required for optimal NOD2-induced IRF5 nuclear translocation (Fig. S5B). Furthermore, IRAK1 and TRAF6, molecules downstream of NOD2 which regulated IRF5-dependent MAPK and NFκB pathway activation (Fig. 5), were also required for IRF5 phosphorylation following NOD2 stimulation (Fig. 6A). Given our previous finding that IRF5 is required for PRR-induced NFκB activation in MDMs (Hedl and Abraham, 2012), we asked if IKKβ and IRF5 cross-regulate phosphorylation of each other. NOD2-induced IKKβ phosphorylation in fact depended on IRF5; as expected IKKβ phosphorylation also depended on NOD2, RIP2, IRAK1, and TRAF6 with MDP treatment (Fig. 6B). Taken together, in addition to IKKβ, RIP2, IRAK1 and TRAF6 are required for NOD2-induced IRF5 phosphorylation, and IRF5, in turn, regulates IKKβ activation.
Fig. 6. RIP2, IRAK1 and TRAF6 are required for IRF5 serine phosphorylation and associate with IRF5 in NOD2-stimulated MDMs.
(A) MDMs were transfected with the indicated siRNA and treated with 100µg/ml MDP for 15min. IRF5 was immunoprecipitated from cell lysates and IRF5 serine phosphorylation was assessed by western blot. Total IRF5 and GAPDH expression from whole cell lysates (WCL) served as loading controls. (B) MDMs (n=8 donors) were transfected with the indicated siRNA. Cells were then treated for 15min with 100µg/ml MDP. Summary graph with fold phospho-IKKβ induction by flow cytometry compared to untreated, scrambled siRNA-transfected cells for each donor+SEM. (C) MDMs were treated with 100µg/ml MDP for 15min. IRF5 was immunoprecipitated from cell lysates and association of the indicated proteins was assessed by western blot. Expression of the respective proteins in whole cell lysates served as a control. IgG isotypes were included as controls (data not shown). Tx, treatment; scr, scrambled. ††, p<1×10−5.
We next sought to better define mechanisms through which IRF5 regulates NOD2-induced signaling. IRF5 has been found to associate with IRAK1 and TRAF6 in overexpression systems and mouse macrophages (Balkhi et al., 2008; Inoue et al., 2014; Takaoka et al., 2005); it is unclear if this is the case in primary human macrophages. Furthermore, association of IRF5 and RIP2 has not been reported. We found that IRF5 associated with RIP2, IRAK1 and TRAF6 in NOD2-stimulated human MDMs (Fig. 6C), consistent with the requirement for each of these molecules in regulating IRF5 phosphorylation following NOD2 stimulation (Fig. 6A). Taken together, IRF5 associates with PRR-signaling intermediates following PRR stimulation, and these intermediates are required for IRF5 phosphorylation and select IRF5-dependent downstream outcomes.
Pro-inflammatory cytokines and glycolysis cross-regulate each other
As the levels of pro-inflammatory cytokines (Fig. 4A) and of glycolysis (Fig. 3C) are high in M1 macrophages, we investigated the relationship between these two PRR-induced outcomes. Glycolytic gene expression and glycolysis were induced by pro-inflammatory cytokines and IRF5 was required for optimal regulation of these outcomes (Fig. S6A&B). Furthermore, blockade of NOD2-induced autocrine pro-inflammatory cytokines in both M0 and M1 macrophages resulted in decreased glycolysis (Fig. S6C), highlighting a role for autocrine pro-inflammatory cytokines in the enhanced glycolysis observed with PRR stimulation. Moreover, knockdown of glycolytic genes (Fig. S6D) and hexokinase inhibition (data not shown) reduced NOD2-induced cytokines, thereby establishing a clear role for glycolysis in PRR-induced cytokine secretion. Taken together, pro-inflammatory cytokines regulate glycolytic outcomes in an IRF5-dependent manner, and pro-inflammatory cytokines and glycolysis cross-regulate each other.
Macrophages from IRF5 Disease-Risk Variant Carriers show increased IRF5 protein expression, increased NOD2-induced Akt2 activation and glycolysis, and increased M1 polarization
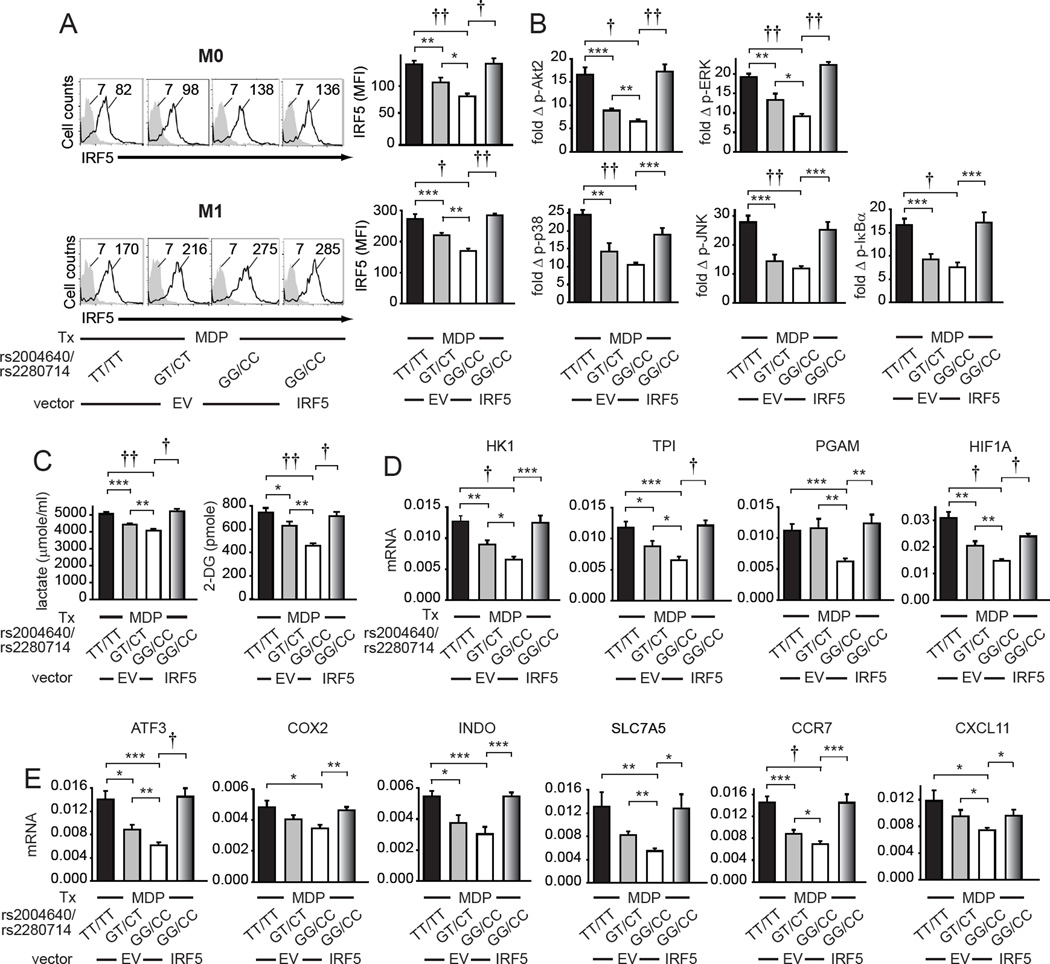
Given the association of IRF5 polymorphisms with multiple immune-mediated diseases (Graham et al., 2006), and the common distribution of the distinct IRF5 genotypes across the population, we sought to define how these variants regulate the IRF5-dependent immune outcomes we defined in this study. Disease-associated rs2004640/rs2280714 TT/TT carriers express increased IRF5 transcript levels relative to GG/CC genotype carriers (Graham et al., 2006), and we previously found that the rs2004640 T allele carriers express increased NOD2-induced cytokines in a gene dose-dependent manner (Hedl and Abraham, 2012). We ensured that following NOD2 stimulation these risk carrier MDMs also expressed increased IRF5 protein relative to non-risk carrier MDMs (Fig. 7A); IRF5 expression was higher in M1 macrophages (Fig. 7A). Heterozygotes showed an intermediate IRF5 expression (Fig. 7A). In M1 macrophages from these disease-risk carriers, NOD2-induced Akt2 activation and MAPK and NFκB pathway activation was increased relative to GG/CC genotype carriers (Fig. 7B). Consistent with the increased Akt2 activation in MDMs from rs2004640/rs2280714 TT/TT disease risk carriers (Fig. 7B), NOD2-induced expression of glycolysis (Fig. 7C), glycolytic pathway genes (Fig. 7D) and HIF1A (Fig. 7D) was increased in M1 macrophages from rs2004640/rs2280714 TT/TT disease risk carriers relative to GG/CC carriers. Further, M1 macrophages from IRF5 risk-allele carriers demonstrated enhanced M1 polarization markers (Fig. 7E). To establish that the higher IRF5 expression in disease-risk carrier MDMs is sufficient to regulate these various critical macrophages outcomes, we transfected the lower IRF5-expressing non-risk carrier rs2004640/rs2280714 GG/CC NOD2-stimulated M1 macrophages with IRF5 to approximate the levels in rs2004640/rs2280714 TT/TT risk carriers (Fig. 7A).
Fig. 7. Macrophages from rs2004640/rs2280714 TT/TT disease-risk carriers show increased IRF5 expression and increased PRR-induced Akt2, MAPK and NFκB pathway activation, glycolysis and M1 polarization.
MDMs from rs2004640/rs2280714 TT/TT, GT/CT or GG/CC genotypes were transfected with empty vector (EV) or an IRF5-expressing vector as indicated for 24h. (A) Cells (n=9/genotype) were left non-polarized (M0) or polarized to M1 macrophages for 24h and then treated for 24h with 100µg/ml MDP. Representative and summarized flow cytometry data for IRF5 protein expression+SEM. Isotype controls (gray shading). (B) M1 macrophages (n=9/genotype) were treated with 100µg/ml MDP for 15min. Summarized flow cytometry data for fold phospho-Akt2, -ERK, -p38, -JNK or -IκBα normalized to untreated cells+SEM. (C) M1 macrophages (n=9/genotype) were treated with 100µg/ml MDP for 24h. Lactate production and glucose uptake. Mean+SEM. (D–E) M1 macrophages (n=9/genotype) were treated with 100µg/ml MDP for 4h. mRNA expression (change in CT values normalized to β-actin and represented as a linear scale)+SEM. Tx, treatment. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5.
NOD2-induced Akt2 activation, glycolysis, glycolytic pathway gene expression, and M1 polarization markers were increased under these conditions (Fig. 7). Taken together, consistent with the increased IRF5 expression in MDMs from IRF5 disease-associated rs2004640/rs2280714 TT/TT carriers, M1 macrophages from these individuals show increased Akt2 activation, glycolytic pathway gene expression, glycolysis, and M1 polarization.
Discussion
We identify distinct IRF5-dependent signaling pathways and mechanisms regulating diverse critical immune outcomes in PRR-stimulated human macrophages. We establish that IRF5-dependent activation of Akt2 signaling is required for expression of glycolytic genes and HIF1A, and in turn, both basal and PRR-induced glycolytic flux. We further find that IRF5 is required for PRR-enhanced glycolysis in mice in vivo. Moreover, pro-inflammatory cytokines and PRR-induced autocrine cytokines regulate glycolysis in an IRF5-dependent manner. IRF5-dependent Akt2 signaling is also necessary for M1 polarization, the enhanced glycolysis in M1 macrophages, and the pro-inflammatory, but not anti-inflammatory cytokine secretion in M0 and M1 macrophages. In contrast, IRF5-dependent MAPK and NFκB pathways contribute to optimal NOD2-induced pro-inflammatory and anti-inflammatory cytokines. The upstream regulation for each of these pathways is also distinct, with BCAP upstream of Akt2, and IRAK1/TRAF6 upstream of MAPK and NFκB. Moreover, IRF5 associates with RIP2, IRAK1 and TRAF6 and each of these molecules, in addition to IKKβ, is required for NOD2-induced IRF5 serine phosphorylation. Thus, in addition to its role as a transcription factor, IRF5 associates with proximal signaling molecules that regulate downstream outcomes as we found in this study, and as has been observed in additional reports (Balkhi et al., 2008; Inoue et al., 2014; Takaoka et al., 2005). Importantly, MDMs from rs2004640/rs2280714 TT/TT IRF5 disease risk carriers show increased NOD2-induced Akt2 activation, glycolysis, M1 polarization, and M1-associated pro-inflammatory cytokines relative to GG/CC carriers. Increasing IRF5 expression in the lower IRF5-expressing rs2004640/rs2280714 GG/CC non-risk allele carrier macrophages increases these outcomes. Taken together, we identify IRF5 as a central regulator of glycolysis and M1 polarization through Akt2 activation, and establish that IRF5 disease-risk carrier macrophages show an increase in these inflammatory outcomes (Fig. S7).
Different Akt isoforms regulate distinct downstream outcomes, including in metabolism, cell proliferation, motility (Hers et al., 2011), and macrophage polarization (Arranz et al., 2012). Therefore, it is crucial to understand how these isoforms are regulated. Until recently studies utilized a phospho-antibody recognizing a motif common to Akt1 and Akt2, such that differential activation was not generally dissected. Previously we used this common phospho-Akt antibody and found that with IRF5 knockdown Akt signaling was intact (Hedl and Abraham, 2012). However, antibodies distinguishing Akt1 and Akt2 phosphorylation are now available and have been successfully used by others (Jeffery et al., 2015). Using these selective antibodies, we now identify that following NOD2 stimulation, while Akt1 phosphorylation remains intact following IRF5 knockdown, Akt2 phosphorylation decreases (Fig. 1). Selectivity in upstream Akt1 and Akt2 activation has not been well-defined; PHLPP1 and PHLPP2 are one such example of molecules targeting specific Akt isoforms (Brognard et al., 2007). Another branch point downstream of NOD2 is the differential signaling intermediates required for activation of the Akt2 pathway and the MAPK and NFκB pathways (Fig. 5). Furthermore, expression of constructs leading to constitutive activation of these pathways is sufficient to partially rescue select decreased outcomes in NOD2-stimulated, IRF5-deficient human macrophages (Figs. 3&4). Of note is that in NOD2-stimulated human macrophages in which MAPKs and NFκB are regulated physiologically, these pathways can play a non-redundant role in PRR-induced cytokines, such that expression of constitutively active signaling molecules may compensate for deficiencies in other pathways. Another interesting observation is that IRAK1 and TRAF6 are required for IRF5 phosphorylation (Fig. 6A), but are not required for Akt2 phosphorylation (Fig. 5A). In contrast, IRF5 is required for Akt2 phosphorylation (Fig. 1F). This suggests that some IRF5 downstream effects depend on IRF5 phosphorylation, at least as detected by the anti-phospho-serine antibody utilized, while others do not. It is possible that different post-translational IRF5 modifications may regulate distinct IRF5-dependent functions. For example, IRF5 phosphorylation is required for nuclear translocation, transcription, and apoptosis, but not for IRF5 ubiquitination (Chang Foreman et al., 2012), such that IRF5 ubiquitination may regulate additional IRF5-dependent downstream events.
Although the association of glycolysis with inflammation in myeloid cells and with inflammatory diseases in general has been well established (Cramer et al., 2003; Krawczyk et al., 2010), mechanisms regulating increased glycolysis following PRR stimulation and in M1 macrophages are unclear, particularly in primary human cells. Mechanisms include increased TLR-induced association of HK2 with mitochondria in mouse DCs (Everts et al., 2012), and increased HK2 and lactate transporter expression in mouse macrophages (Tan et al., 2015). We find that NOD2 stimulation increases glycolysis by upregulating HK1, PGAM, TPI, and HIF1A in an IRF5-dependent manner (Fig. 1); IRF5 also regulates basal expression of multiple additional glycolytic genes (Fig. 1). To our knowledge, PRR-mediated regulation of PGAM and TPI, and NOD2- and Akt2-mediated regulation of glycolytic genes in general has not been reported. Mutations in glycolytic genes can regulate immune-mediated and infectious disease susceptibility (Ayi et al., 2008). While the contribution of glycolysis to immune-mediated diseases has been mostly studied in the context of T cells (Ganeshan and Chawla, 2014), studies also show an important role for glycolysis in myeloid cells in immune-mediated diseases. For example, loss of the glycolytic regulator HIF-1α decreases experimental arthritis and cutaneous inflammation in mice through regulating myeloid cell contributions to these diseases (Cramer et al., 2003). We now find that HIF1A is regulated in human MDMs upon PRR stimulation in a manner dependent on disease-associated IRF5 polymorphisms (Fig. 7). We further identify that these common IRF5 polymorphisms associated with multiple immune-mediated diseases can modulate glycolysis in general.
This study identifies IRF5 as a critical link that integrates PRR-induced and M1-enhanced glycolysis and M1 polarization through Akt2 signaling, thereby highlighting key contributions and mechanisms of IRF5 to immune outcomes. Furthermore, we establish that macrophages from carriers of IRF5 variants associated with increased IRF5 protein expression and multiple immune-mediated diseases show increased PRR-induced Akt2 activation, glycolysis and M1 polarization. IRF5 polymorphisms are commonly distributed across the population and contribute to susceptibility to multiple immune-mediated diseases, making IRF5 a promising target for therapy of such diseases. Our studies indicate that IRF5 might have additional roles in multiple biological pathways regulated by glycolysis, and we identify mechanisms and pathways differentially regulated by IRF5 that could be targeted to decrease undesired inflammation.
Experimental Procedures
Patient recruitment and genotyping
Informed consent was obtained per protocol approved by the institutional review board at Yale University. We recruited healthy individuals with no personal or family history of autoimmune/inflammatory disease, including psoriasis, SLE, rheumatoid arthritis, multiple sclerosis, type I diabetes mellitus, Crohn’s disease, and ulcerative colitis, or a history of HIV. Genotyping was performed by Taqman (Life Technologies, Grand Island, NY) or Sequenom Platform (Sequenom, San Diego, CA).
Primary myeloid cell culture and MDM polarization
Human peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood using Ficoll-Paque (GE Dharmacon, Lafayette, CO). Monocytes were purified from PBMCs by positive CD14 selection (Miltenyi Biotec, Auburn, CA) or adhesion (purity was >98% by CD11c expression), and cultured in RPMI 1640 medium pH 7.3 containing 100 U/ml penicillin and 100 µg/ml streptomycin, 1 mM sodium pyruvate, MEM non-essential amino acids, 10 mM HEPES (Thermo Fisher Scientific, Waltham, MA), 10% fetal bovine serum and M-CSF (10ng/ml) (Shenandoah Technology, Warwick, PA) for 7 days for MDM differentiation. In some studies, MDMs were treated for 24h with 100ng/ml LPS (Sigma-Aldrich, St. Louis, MO) and 20ng/ml IFN-γ (R&D Systems, Minneapolis, MI) (M1 polarization) or 20ng/ml IL-4 (R&D Systems) (M2 polarization).
Mice
IRF5−/− mice (C57BL/6) were generously provided by Dr. Betsy J. Barnes. Mice were backcrossed with C57BL/6 mice to generate littermate controls. Mice were maintained on autoclaved food in a specific pathogen-free facility. Experiments were performed in agreement with the Yale University Institutional Animal Care and Use Committee and National Institutes of Health guidelines.
Myeloid cell stimulation
MDMs were treated with MDP (Bachem, Torrance, CA). Supernatants from 3×105 MDMs/well in 24 well plates were assayed for TNF-α (clones MAb1 and Mab11), IL-8 (clones G265-5 and G265-8), IL-10 (clones JES3-9D7 and JES3-12G8) (BD Biosciences), or IL-12p40 (clones C8.3 and C8.6), IL-1Ra (clone CRM17 and #13-7014-81) and IL-1β (clones CRM56 and CRM57) (eBioscience) by ELISA.
Lactate production, glucose uptake and hexokinase assays
Following stimulation of 5×104 MDMs/well in a 96 well plate, conversion of lactate to pyruvate was measured as a change in NAD+ absorbance at 340 nm as per (Sekine et al., 1994), 2-deoxyglucose (VWR) uptake measured as per (Yamamoto et al., 2006) and hexokinase activity per manufacturer’s instruction (Biovision).
Constructions and transfection of siRNA and DNA vectors
100nM scrambled or siGENOME SMARTpool siRNAs (a pool of four distinct, commercially designed siRNA) against IRF5 (#M-011706-00), Akt1 (#M-003000-03), Akt2 (#M-003001-02), NOD2 (#M-003464-01), IKKβ (#M–003503–03), IRAK-1 (#M-004760-03), TRAF6 (#M-004712-00), BCAP (#M-016285-01), RIP2 (#M-003602-02), HK1 (#M-006820-01), TPI1 (#M-009776-02), PGAM (#M-008883-01), or HIF1A (#M-004018-05) (GE Dharmacon) or 4µg pMCL-MKK1 (R4F) (constitutively active ERK kinase)(Mansour et al., 1994), 4µg pSRα-3HA-JNKK2-JNK1-WT (constitutively active JNK)(Zheng et al., 1999) (generous gifts from Dr. Ben Turk), 4µg pcDNA3-Flag MKK6(glu) (constitutively active p38 kinase) (Addgene plasmid 13518; kindly deposited by Roger Davis (Raingeaud et al., 1996)), 2µg IKK-2 S177D S181E (constitutively active NFκB) (Addgene plasmid 11105; kindly deposited by Anjana Rao (Mercurio et al., 1997)), 2 µg pcDNA3 Myr HA Akt2 (constitutively active Akt2) (Addgene plasmid 9016, kindly deposited by William Sellars), 2µg IRF5 vector (Genecopoeia, Rockville, MD) or empty vector (a control vector without the gene of interest) were transfected for 48h (unless otherwise indicated) into MDMs using Amaxa nucleofector technology (Amaxa, San Diego, CA). Transfection efficiency was >70%.
Protein detection
Intracellular proteins were detected in permeabilized cells by flow cytometry with Alexa Fluor 647-, phycoerythrin-, Alexa Fluor 488- or unlabeled antibodies to phospho-ERK (D13.14.4E), phospho-p38 (3D7), phospho-JNK (G9), phospho-IκBα (14D4) (Cell Signaling Technology, Danvers, MA), phospho-Akt1 (#ab81283), phospho-Akt2 (#ab110231) (Abcam, Boston, MA) and IRF5 (1H6) (Novus, Littleton, CO). Briefly, 0.8–1×106 cells were fixed with Lyse/Fix Buffer (BD Biosciences) for 10 min, permeabilized for 1h with Perm Buffer III (BD Biosciences), washed and then stained with the indicated antibodies. RIP2 (25/RIG-G, BD Biosciences), TRAF6 (D21G3) and IRAK-1 (D51G7) (Cell Signaling Technology) expression was detected by western blot and/or flow cytometry. GAPDH antibody (6C5, EMD Millipore, Billerica, MA) was used to assess equal loading. For immunoprecipitation and western blot, 2–5×106 macrophages were lysed and lysates were centrifuged at 10,000g for 10 min at 4°C. For immunoprecipitation anti-IRF5 antibody (E1N9G, Cell Signaling Technology)-bound protein A Sepharose (EMD Millipore) was incubated with pre-cleared lysates at 4°C overnight. Immunoprecipitates were run on 10% SDS-PAGE gels and transferred to PVDF membrane (Bio-Rad, Hercules, CA). IRF5 serine phosphorylation was examined by incubation with an anti-phospho-(Ser) R/KXXpSXP motif antibody (E2, Cell Signaling Technology). Signal was detected by secondary anti-rabbit or anti-mouse IgG DyLight 680 and DyLight 800 antibodies (Thermo Fisher Scientific) with the Odyssey Imager (Li-COR Biosciences, Lincoln, NE). Densitometry was performed using Image J software (NIH).
mRNA expression analysis
Following stimulation, total RNA was isolated from 0.5–1×106 MDMs using Trizol reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. RNA was treated with DNAse (Thermo Fisher Scientific) and reverse transcribed using first strand cDNA synthesis with Superscript III Reverse Transcriptase and Oligo dT (Thermo Fisher Scientific). Quantitative PCR was performed using 100 ng RNA equivalents/sample using All-In-One qPCR Mix (Genecopoeia) as in (Hedl et al., 2007) on the ABI Prism 7000 (Applied Biosystems). Samples were normalized to β-actin. Primer sequences are available upon request.
Statistical analysis
Significance was assessed using a two-tailed Student’s t-test. p<0.05 was considered significant. Lines over adjacent bars indicate identical p values for these bars.
Supplementary Material
Highlights.
IRF5 regulates glycolysis in PRR-stimulated human macrophages in vitro and in mice
IRF5 increases glycolysis through Akt2 activation and glycolytic gene induction
IRF5 associates with RIP2, IRAK1, and TRAF6 in human macrophages upon NOD2 stimulation
IRF5 disease variant cells show increased signaling, glycolysis, and M1 polarization
Acknowledgments
This work was supported by AI110458, AI120369, DK099097, P30-DK34989 from the National Institutes of Health and the Crohn’s and Colitis Foundation of America. We gratefully acknowledge Drs. Betsy Barnes and Ben Turk for reagents.
Abbreviations
- IBD
inflammatory bowel disease
- IRF
interferon-regulatory factor
- MDMs
monocyte-derived macrophages
- MDP
muramyl dipeptide
- NOD2
nucleotide-binding oligomerization domain containing 2
- PRR
pattern-recognition receptor
- SLE
systemic lupus erythematosus
- TLR
Toll–like receptor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
M.H., J.Y. and C.A. designed and conducted the experiments, M.H. and C.A. wrote the paper.
References
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayi K, Min-Oo G, Serghides L, Crockett M, Kirby-Allen M, Quirt I, Gros P, Kain KC. Pyruvate kinase deficiency and malaria. N Engl J Med. 2008;358:1805–1810. doi: 10.1056/NEJMoa072464. [DOI] [PubMed] [Google Scholar]
- Balkhi MY, Fitzgerald KA, Pitha PM. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol Cell Biol. 2008;28:7296–7308. doi: 10.1128/MCB.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22:5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom B, Aune MH, Awuh JA, Kojen JF, Blix KJ, Ryan L, Flo TH, Mollnes TE, Espevik T, Stenvik J. TLR8 Senses Staphylococcus aureus RNA in Human Primary Monocytes and Macrophages and Induces IFN-beta Production via a TAK1-IKKbeta-IRF5 Signaling Pathway. J Immunol. 2015;195:1100–1111. doi: 10.4049/jimmunol.1403176. [DOI] [PubMed] [Google Scholar]
- Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Chang Foreman HC, Van Scoy S, Cheng TF, Reich NC. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One. 2012;7:e33098. doi: 10.1371/journal.pone.0033098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmas E, Toubal A, Alzaid F, Blazek K, Eames HL, Lebozec K, Pini M, Hainault I, Montastier E, Denis RG, et al. Irf5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nat Med. 2015 doi: 10.1038/nm.3829. [DOI] [PubMed] [Google Scholar]
- Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, Pearce EJ. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan-Pena S, O'Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Pons-Estel B, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- Hedl M, Abraham C. IRF5 risk polymorphisms contribute to interindividual variance in pattern recognition receptor-mediated cytokine secretion in human monocyte-derived cells. J Immunol. 2012;188 doi: 10.4049/jimmunol.1103319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Inoue M, Arikawa T, Chen YH, Moriwaki Y, Price M, Brown M, Perfect JR, Shinohara ML. T cells down-regulate macrophage TNF production by IRAK1-mediated IL-10 expression and control innate hyperinflammation. Proc Natl Acad Sci U S A. 2014;111:5295–5300. doi: 10.1073/pnas.1321427111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Church CD, Holtrup B, Colman L, Rodeheffer MS. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol. 2015;17:376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA. IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, Cross JR, Jung E, Thompson CB, Jones RG, Pearce EJ. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook AD, Hamilton JA. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- Lazzari E, Jefferies CA. IRF5-mediated signaling and implications for SLE. Clin Immunol. 2014;153:343–352. doi: 10.1016/j.clim.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Lopez-Pelaez M, Lamont DJ, Peggie M, Shpiro N, Gray NS, Cohen P. Protein kinase IKKbeta-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- Marsin AS, Bouzin C, Bertrand L, Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J Biol Chem. 2002;277:30778–30783. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, MacFarlane AWt, Toft M, Lowell CA, Campbell KS, Hamerman JA. B-cell adaptor for PI3K (BCAP) negatively regulates Toll-like receptor signaling through activation of PI3K. Proc Natl Acad Sci U S A. 2012;109:267–272. doi: 10.1073/pnas.1111957108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Opitz B, Puschel A, Schmeck B, Hocke AC, Rosseau S, Hammerschmidt S, Schumann RR, Suttorp N, Hippenstiel S. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J Biol Chem. 2004;279:36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- Panasyuk G, Espeillac C, Chauvin C, Pradelli LA, Horie Y, Suzuki A, Annicotte JS, Fajas L, Foretz M, Verdeguer F, et al. PPARgamma contributes to PKM2 and HK2 expression in fatty liver. Nat Commun. 2012;3:672. doi: 10.1038/ncomms1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, Sassetti CM, Kelliher MA. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pore N, Liu S, Shu HK, Li B, Haas-Kogan D, Stokoe D, Milanini-Mongiat J, Pages G, O'Rourke DM, Bernhard E, Maity A. Sp1 is involved in Akt-mediated induction of VEGF expression through an HIF-1-independent mechanism. Mol Biol Cell. 2004;15:4841–4853. doi: 10.1091/mbc.E04-05-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Chen X, Chen ZJ. IKKbeta is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci U S A. 2014;111:17438–17443. doi: 10.1073/pnas.1418516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieske P, Pongubala JM. AKT induces transcriptional activity of PU.1 through phosphorylation-mediated modifications within its transactivation domain. J Biol Chem. 2001;276:8460–8468. doi: 10.1074/jbc.M007482200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, Bosca L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba DG, Heger A, Eames HL, Oikonomopoulos S, Teixeira A, Blazek K, Androulidaki A, Wong D, Goh FG, Weiss M, et al. IRF5:RelA interaction targets inflammatory genes in macrophages. Cell Rep. 2014;8:1308–1317. doi: 10.1016/j.celrep.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine N, Cirulli V, Regazzi R, Brown LJ, Gine E, Tamarit-Rodriguez J, Girotti M, Marie S, MacDonald MJ, Wollheim CB, et al. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. J Biol Chem. 1994;269:4895–4902. [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- Tan Z, Xie N, Banerjee S, Cui H, Fu M, Thannickal VJ, Liu G. The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J Biol Chem. 2015;290:46–55. doi: 10.1074/jbc.M114.603589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutman TD, Hu W, Fulenchek S, Yamazaki T, Kurosaki T, Bazan JF, Pasare C. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc Natl Acad Sci U S A. 2012;109:273–278. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sandling JK, Hagberg N, Berggren O, Sigurdsson S, Karlberg O, Ronnblom L, Eloranta ML, Syvanen AC. Genome-wide profiling of target genes for the systemic lupus erythematosus-associated transcription factors IRF5 and STAT4. Ann Rheum Dis. 2013;72:96–103. doi: 10.1136/annrheumdis-2012-201364. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Sato T, Kawasaki K, Murosaki S, Yamamoto Y. A nonradioisotope, enzymatic assay for 2-deoxyglucose uptake in L6 skeletal muscle cells cultured in a 96-well microplate. Anal Biochem. 2006;351:139–145. doi: 10.1016/j.ab.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Zheng C, Xiang J, Hunter T, Lin A. The JNKK2-JNK1 fusion protein acts as a constitutively active c-Jun kinase that stimulates c-Jun transcription activity. J Biol Chem. 1999;274:28966–28971. doi: 10.1074/jbc.274.41.28966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.