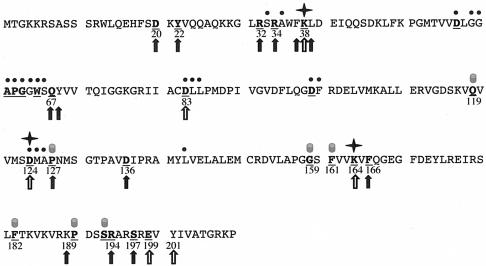
FIG. 1.
Rational mutant design. The 27 most highly conserved residues in RrmJ are shown in bold and underlined letters. These amino acids have been identified as highly conserved by aligning a set of 1,112 nonredundant paralog sequences to a set of 29 likely RrmJ orthologs (6). The black dots above the RrmJ sequence indicate the residues that have been identified as involved in AdoMet binding (6). Stars mark the three amino acids that have been shown to form the catalytic triad (16). The filled cylinders above the sequence depict the amino acids which are either predicted to play a structural role in RrmJ or reside on the opposite site of the RrmJ molecule. The open arrows mark all highly conserved residues that were mutated in previous studies (16). The closed arrows depict the residues that were mutated in this study.