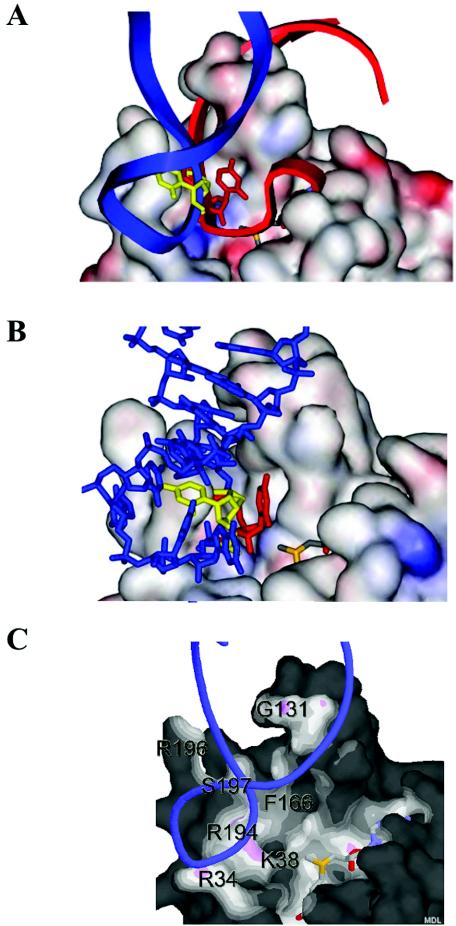
FIG. 5.
Modeling of the A loop onto the surface of RrmJ. (A) In situ modeling of the A-loop structure (nucleotides 2548 to 2560) as solved by NMR (3) onto the surface of RrmJ. Modeling is based on overlaying the U2552 reactive nucleotide into the active site of RrmJ with the 5′ m7G-capped reactive nucleotide in the VP39 structure. Without turning the A loop, the double helical stalk of the A loop clashes severely with the extended α4 helix of RrmJ (red model). To avoid this clashing, the A loop is turned by 85 degrees (blue model). Now, phosphodiesters occupy the same location in the model of RrmJ as do phosphodiesters of the 5′ m7G-capped RNA hexamer in the VP39 structure. (B) Base flipping of U2552. The model shows U2552 in the closed position (yellow), where the 2′ hydroxyl group of the ribose is not accessible to the methyl donor AdoMet, and in the flipped configuration (red), where the 2′ hydroxyl group is in the optimal position to be methylated. (C) Solvent-accessible surface of RrmJ colored by distance to ligand atoms from U2552-flipped A-loop RNA (blue ribbon) and AdoMet (shown as stick model). The surface is colored by distance from nearby atoms. Dark areas are too far away from any atoms outside the surface to be bonded. White or light areas are close enough for hydrophobic van der Waals interactions. Pink areas are close enough for hydrogen bonds. The figure was made with Protein Explorer (25).