Abstract
Viral latency can be considered a metastable, nonproductive infection state that is capable of subsequent reactivation to repeat the infection cycle. Viral latent infections have numerous associated pathologies, including cancer, birth defects, neuropathy, cardiovascular disease, chronic inflammation, and immunological dysfunctions. The mechanisms controlling the establishment, maintenance, and reactivation from latency are complex and diversified among virus families, species, and strains. Yet, as examined in this review, common properties of latent viral infections can be defined. Eradicating latent virus has become an important but elusive challenge and will require a more complete understanding of the mechanisms controlling these processes.
Introduction
If Tolstoy were a virologist, he may have concluded that “lytic viruses are all alike; every latent virus is latent in its own way.” And while each virus may be unique, there are common themes governing viral latent infection. Viral latency can best be defined as (1) a long-term persistent infection (2) with reservoirs that fail to produce infectious virus and (3) that are capable of reactivating to repeat the infection cycle, a process referred to as reversibility (Speck and Ganem, 2010). Latency has also been described as “long-term parking” (Flint et al., 2015) and a “round trip” infection (Klein, 1976), which help to describe the distinct life cycle events of establishing, maintaining, and reactivating from latency. Viral latent infections are neither innocuous, nor inert. Rather, they are highly dynamic interactions in metastable equilibrium with host antiviral immunity. Chronic infection and immunological disequilibrium can lead to devastating disease, including cancer and autoimmune disorders. Eradicating latent reservoirs has become an important but elusive challenge for treatment of human immunodeficiency virus (HIV), human papillomavirus (HPV), hepatitis B virus (HBV), and herpesviruses. Therefore, a better understanding of the molecular and cellular processes controlling viral latency will be required to address this challenging unmet biomedical concern.
General Themes of Viral Latency
Viruses frequently fail to complete their life cycle. Nonproductive infection can be a result of many factors, including host cell restriction factors, inadequate host resources, defective viral genome replication, or programmed delays in the infection cycle. Although latency can be distinguished from abortive lytic infection, the same factors that restrict and abort productive infection can contribute positively and essentially to the formation of latency (Figure 1A). Different types of latency can also exist, depending on the host cell-type and specific virus or strain. Latency can range from selective viral gene expression with partial replication to a complete quiescence with no detectable viral gene expression or replication. An important question is whether latency is a defined genetic program, or an epiphenomena of a stuttering infection.
Figure 1. Overview of the Dynamic Host-Pathogen Conflict Driving Viral Latency.
(A) Viruses exploit and reprogram host antiviral resistances to adopt a stable latent infection.
(B) Host intrinsic resistances that restrict productive lytic cycle infection can drive the formation of latency. DNA-damage sensing and chromatin assembly factors that limit productive infection can promote latent episomes and stable chromatin formation. Latency-associated noncoding RNA evade immune detection and reroute inflammatory pathways to promote conditions permissive for latent infection (see text for details).
The genetic factors regulating viral infection are known to have accelerated rates of evolution due to the “arms race” between host and pathogen (Daugherty and Malik, 2012). This dynamic is especially important for genes controlling viral latency, which is a long-term détente between virus and host. For many viruses, genes involved in the regulation of latency have faster rates of divergence relative to the more conserved genes encoding viral structural proteins and replication enzymes. Viral latency commonly involves the coopting of host intrinsic defenses to promote viral persistence (Figure 1B). As may be expected, this repurposing is most commonly observed in immune cells (hematopoietic lineage), or immune privileged cells (neuronal, postmitotic cells), where the virus may find a long-term “safe house” to convert an antiviral enemy into an asset for latency.
Temporal control of viral gene expression is integrally linked with viral latency. Herpesvirus genes have a tightly controlled temporal order of expression mechanistically determined by a dependence on preceding events, like DNA replication preceding late gene transcription. The establishment of latency and reactivation from latency may be determined, in part, from variations in the temporal program of a typical productive infection. There are examples of aberrant or alternative gene programs, including partial or poised lytic infection, with blocks to virus production occurring at various stages of the temporal cascade. This can reflect host cell type restrictions, as well as virus subtypes (e.g., HPV), that can determine whether a stable latent infection will be established.
Latency Reservoirs and Reactivation Rates in Natural Infection
Virus latency is a dynamic process at the cellular and organismal level. At the organismal level, the site and size of latent reservoirs can determine the stability of latency and frequency of reactivation. The extent of latent infection cannot always be measured directly, especially in humans. The latent reservoirs of HIV has been estimated to be between 1 and 6 × 106 CD4+ T cells based on the levels and rates of virus detected after disruption of antiviral therapy (Pinkevych et al., 2015). And the tissue distribution of these T cells may be equally important for stable latency. Similarly, herpes simplex 2 (HSV2) shedding can be detected continuously and chronically in subclinical latent infections, suggesting that latency is multifocal and subject to chronic reactivation (Johnston and Corey, 2016). Epstein-Barr virus (EBV), which establishes an asymptomatic long-term latent infection in 90% of healthy adults, is detected chronically in saliva and blood at variable levels. Mathematical modeling predicts that stable latent infection of EBV requires a dynamic equilibrium between reactivation and reinfection (Thorley-Lawson, 2015). The equilibrium rate of reactivation is estimated to be 1–3 reactivating patches in the oropharyngeal lymphoid tissue, with each reactivating field containing between 1 and 105 lytic infected epithelial cells (Hadinoto et al., 2009). Many of these reactivation events are predicted to be abortive, or nonproductive. Thus, organismal latency is a highly dynamic process that involves chronic reactivation and re-establishment of latent reservoirs at the cellular level.
Genetic Diversity: Strains and Polymorphisms
Viral strains and polymorphisms are known to have significant effects on the overall biology of infection and its pathogenesis. However, it is not known to what extent virus sequence variation contributes to latency. The variety of high- and low-risk subtypes of HPV has a well-established contribution to carcinogenesis, but it is less clear how these subtypes correspond to persistence, latency, and reactivation of HPV. Laboratory strains of human cytomegalovirus virus (HCMV) have lost substantial regions of their genomes, which are now known to contain genes important for the establishment of latent infection in vivo (Murphy and Shenk, 2008). While the loss of these genes may be due to artificial cell culture selection pressure, it is not yet known whether mutations and polymorphisms in these genes contribute to latency decisions and different clinical outcomes. Next-generation sequencing has revealed large numbers of polymorphisms in the latency-associated genes of EBV (Palser et al., 2015). Notably, a single amino acid polymorphism in EBNA2 confers more efficient immortalization of primary B-lymphocytes (Farrell, 2015), variants in LMP1 are enriched in nasopharyngeal carcinoma (NPC) (Edwards et al., 2004), deletions of EBNA2 are found in Burkitt’s lymphoma (BL) (Kelly et al., 2009), mutations in EBNA3B in Diffuse Large B cell Lymphoma (DLBCL) (White et al., 2012), and mutations in RPMS1 protein correlate with increase NPC risk (Feng et al., 2015). Viruses derived from NPC were also found to have mutations that increased spontaneous lytic reactivity and epithelial cell tropism (Tsai et al., 2013). These findings emphasize the importance of viral variants that may contribute to latency control and associated pathogenesis.
Tissue Tropism for Latent Infection
Most viruses establish latency only in specialized host cell types and microenvironments that provide essential conditions enabling viral latency. For example, the predominant latent forms of EBV are found in CD19+ memory B cells, HCMV in CD34+ myeloid progenitor cells, and HPV in basal epithelial cells. However, some viruses can establish latency in multiple cell types. HCMV latent infection is also found in endothelial cells and macrophages (Fish et al., 1995), neuronal progenitors (Belzile et al., 2014) and myeloid precursor cells (Taylor-Wiedeman et al., 1994). EBV latent infection can be found in epithelial cells (Tsao et al., 2015), but these are typically associated with cancer phenotypes, and are thought to be aberrations in the normal viral life cycle (Moore and Chang, 2010). Latency typically occurs in cells that are nonpermissive for lytic cycle gene expression (reviewed in Poole and Sinclair, 2015). But latency may also require cellular and tissue microenvironments that are uniquely permissive for latent cycle gene expression. In many cases, the permissive cell type and microenvironment cannot be recapitulated in tissue culture. For example, most EBV-positive NPC cells or Kaposi’s sarcoma-associated herpesvirus (KSHV)-positive Kaposi’s sarcoma (KS) cells lose viral genomes when cultured ex vivo (Dictor et al., 1996; Gullo et al., 2008). Presumably, tissue microenvironment plays a critical role in maintaining these forms of latent infection. Complex tissue culture models are required to recapitulate some aspects of viral latency. For example, HSV1 latency can be modeled in primary rat neurons treated with nerve growth factor (NGF), which maintains progenitor neuronal cell survival and prevents viral lytic cycle gene expression (Camarena et al., 2010; Wilcox and Smith, 1998).
Genome Maintenance Mechanisms
Latent viruses have evolved several mechanisms to maintain viral genome integrity (Figure 2). For many herpesviruses, DNA end protection during latency is achieved by forming “endless” genomes, which typically correspond to closed circular, nonintegrated plasmids, referred to as episomes. Episomes are also found in HPV and HBV latency. However, some herpesviruses integrate into the host chromosome during latency, resembling HIV and retrovirus integration, but achieving this through homologous recombination rather than with viral-encoded integrase.
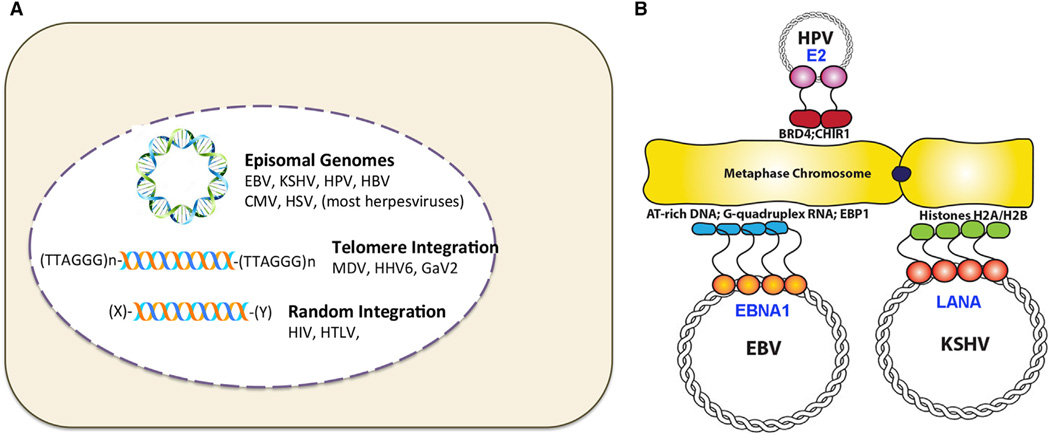
Figure 2. Comparing Latency Origins and Episome Maintenance Elements from Different Viruses.
(A) Episomal genomes for EBV, KSHV, HPV, HBV, CMV, and HSV have been identified during latent infection. Genome integration at telomeres are found forMDV, HHV6, and GaV2 latency, while retroviruses, HTLV, and HIV integrate more randomly.
(B) Episome maintenance factors for HPV E2, EBV EBNA1, and KSHV LANA and their related mechanisms of metaphase chromosome tethering.
Episome Maintenance Factors
Viruses that establish latent infection in proliferating cells require a mechanism to faithfully transmit their genomes during cellular division. Since viral replication proteins are not expressed in latency, these viruses depend upon host cell replication machinery. For integrated viruses, like retroviruses, this is solved by replicating as an integral part of the host genome. However, for nonintegrating episomal genomes, like EBV, KSHV, and HPV, the virus must recruit the host cell replication machinery to the episome and maintain a stable episome copy number by segregating newly replicated genomes equally to daughter cells after each cell division. These viruses encode a structurally-related sequence-specific DNA-binding protein that tethers the viral episome to the host metaphase chromosome during mitosis. EBV EBNA1 (Frappier, 2015), KSHV LANA (Ballestas and Kaye, 2011; Uppal et al., 2014), and HPV E2 (McBride, 2013) share this function. Each protein interacts with host metaphase chromatin through multiple, and distinct cellular targets, including AT-rich DNA for EBNA1, core histones H2A and H2B for LANA, and chromatin associated factors BRD4 and ChlR1 for E2. Whether these different mechanisms of metaphase chromatin attachment have functional significance with respect to viral genome segregation, or whether the tethering by various mechanisms are functionally equivalent, remains to be determined. Regardless of the mechanism, tethering to a metaphase chromosome is essential for episome maintenance and genome persistence in rapidly dividing cells.
Recently, a potential episome maintenance factor has been identified for HCMV latency (Tarrant-Elorza et al., 2014). Pari and colleagues identified a latency associated protein expressed from HCMV IE exon 4 (IE1×4) found in latently infected CD34+ cells. IE1×4 in association with host DNA binding protein Sp1 and topoisomerase IIB binds to the HCMV terminal repeats to facilitate latent cycle DNA replication and episome maintenance (Tarrant-Elorza et al., 2014). While IE1×4 may also stimulate lytic replication, these findings raise the possibility that beta-herpesviruses share common features with the episome maintenance mechanisms used by gammaherpesviruses and HPV.
Genome Integration
Several herpesviruses, like Marek’s Disease virus (MDV), gallid herpesvirus 2 (GaHV-2), and human herpesvirus 6 (HHV6), establish latent infections in CD4+ T cells with viral genomes integrated into host chromosomes (reviewed in Gennart et al., 2015). These viruses frequently integrate into telomere repeat tracts by homologous recombination (Kaufer et al., 2011; Robinson et al., 2010). The viral genome terminal repeats contain stretches of cellular telomere TTAGGG repeats. Precisely why these viruses have captured cellular telomere repeats and use these chromosomal sites for integration during latency is not known, but it is possible that telomeres provide a unique recombinogenic environment conducive for viral integration and mobilization during reactivation. For MDV, telomere integration enhances tumor formation (Greco et al., 2014). It is not yet clear whether these integrated viral genomes can reactivate efficiently to complete their viral productive cycles or if these integrations are pathogenic aberrations.
Epigenetics and Viral Chromatin
DNA in the nucleus must be assembled into some form of nucleoprotein structure to avoid DNA damage signaling and nucleolytic attack. Nucleosomes and chromatin form various structures on cellular DNA, and varieties of chromatin are typically found on latent viral genomes, in formations similar, if not indistinguishable from host cellular DNA. Viral genomes need chromatin for protection and to execute their gene expression programs correctly. However, chromatin can also be repressive, and certain types of heterochromatin may serve as a restriction factor for invading viruses and endogenous retrotransposons. The assembly and distribution of active, euchromatic chromatin or repressive, heterochromatin on viral genome can determine the fate of the virus, including the ability to establish latent infection.
Chromatin Assembly
The nuclear domain 10/PML-associated nuclear bodies are dynamic subnuclear structures that colocalize with viral genomes at early stages of nuclear infection (reviewed in Everett and Chelbi-Alix, 2007; Maul, 1998). The major components of the ND10s are PML, SP100, Daxx, and ATRX, which have all been implicated in antiviral functions. Daxx and ATRX assemble histone H3.3 associated heterochromatin on newly infecting viral genomes (reviewed in Tsai et al., 2015). Viruses encode various tegument proteins that can either degrade or modulate these chromatin assembly factors to regulate lytic or latent gene expression programs. Among the best characterized is the HSV-1 encoded ICP0 protein shown to degrade PML and SP100 through a ubiquitin and SUMO-dependent pathway (reviewed in Boutell and Everett, 2013). The HCMV tegument protein pp71 degrades DAXX, preventing its repression of the major immediate early promoter (MIEP) region during primary infection and reactivation (Saffert and Kalejta, 2006, 2007). Gammaherpesvirus tegument proteins target various components of ND10, and the different interactions may reflect different propensities toward latency (Tsai et al., 2015). For EBV, the tegument protein BNRF1 prevents ATRX-dependent repression of viral latency transcripts, but may not block Daxx repressive chromatin assembly at other sites on the viral genome to prevent lytic gene expression (Tsai et al., 2014). According to this model, selective control of chromatin assembly can promote latency (Figure 1B).
Histone Modifications
Histone tail modifications determine the nature of the chromatin assembled on viral genomes. Studies with HSV1 reveal that histone modification during infection is a highly dynamic process (Kristie, 2015). The cellular chromodomain protein CHD3 recognizes and remodels H3K9me3 and H3K27me3 heterochromatin on the bulk viral genome (Arbuckle and Kristie, 2014). HSV1 tegument protein VP16 selectively reverses viral-associated heterochromatin through interaction with host factors OCT1, a sequence-specific DNA binding factor, and Host-Cell Factor 1 (HCF1), a transcriptional coregulator, to recruit histone demethylases JMJD2s and LSD1 to viral immediate early genes. This allows histone methylases SETDB1 and MLL family members to generate euchromatic H3K4me3 necessary for RNA polymerase II transcription initiation. During latency in neurons, HCF1 and VP16 are excluded from the nucleus, and the Co-REST complex, a transcriptional corepressor complex, inhibits lytic cycle gene expression by recruiting histone deacetylases (reviewed in Roizman et al., 2011). The latent HSV1 genome is enriched with heterochromatic H3K27me3 and H3K9me3, except at the transcriptionally active LAT locus, which has euchromatic marks. Silencing during latency appears to occur in two stages, with CoREST complex functioning at the earliest stages (Du et al., 2010), and Polycomb-mediated H3K27me3 occurring at a later stage (Cliffe et al., 2013).
During HCMV latency, MIEP assembles into repressive chromatin through a process dependent on the CMV UL138 gene (Lee et al., 2015b; Poole and Sinclair, 2015). UL138 was shown to inhibit MIEP recruitment of histone demethylases (KDMs) JMJD3 (KDM6B) and LSD1 (KDM1A) that demethylate and reverse heterochromatic H3K27me3 and K3K9me2, respectively.
Hepatitis B (HBV) covalently closed circular DNA (cccDNA) is also organized into a chromatin-like structure with histones carrying epigenetic marks (Bock et al., 2001; Tropberger et al., 2015). ChIP-Seq revealed that euchromatic histone H3K4me3 was elevated at genome positions corresponding to latency transcript start sites and enhancer like elements (Tropberger et al., 2015). HBV genomes lacking the viral encoded protein HBx are transcriptionally silenced with an increase in SETDB1-dependent histone H3K9 tri-methylation and heterochromatin proteins 1 (HP1) (Rivière et al., 2015). HBx was also found to antagonize Spindlin, a Tudor-domain protein implicated in binding methylated histones (Ducroux et al., 2014). Spindlin was found to inhibit HBV, as well as HSV1 gene expression, suggesting it is a general inhibitor of nuclear DNA viruses.
Polycomb Repression, H3K27me3
The Polycomb complex is associated with facultative heterochromatin and has been implicated in the control of latency for numerous viruses. Polycomb-mediated repression maintains latency for KSHV (Günther and Grundhoff, 2010; Toth et al., 2013; Toth et al., 2010), HSV1 (Cliffe et al., 2013; Watson et al., 2013), HCMV (Abraham and Kulesza, 2013), EBV (Allday, 2013), HPV (Hyland et al., 2011), HBV (Andrisani, 2013), and HIV(Friedman et al., 2011; Matsuda et al., 2015). In some cases, viral encoded oncoproteins (like EBV EBNA3C and HBV pX) direct Polycomb proteins to silence cellular tumor suppressors to help maintain latency by immortalizing the host cell. In other cases, like KSHV and HSV1, Polycomb factors are recruited to the viral genomes to silence lytic genes during latent infection. It is not clear whether the silencing of host and viral genes by Polycomb occurs through the same mechanisms. Polycomb silencing is often mediated by noncoding RNAs. KSHV PAN RNA has been shown to mediate the Polycomb-dependent silencing of host inflammatory cytokines, as well as the KSHV lytic origin (Rossetto and Pari, 2014; Rossetto et al., 2013b). HSV-1 LAT (see below) has also been implicated in recruiting Polycomb factors to repress viral lytic genes (Cliffe et al., 2013).
TRIM28/KAP1
The nuclear protein KAP1 has been shown to play an important role in the epigenetic repression of endogenous retroviruses (Rowe et al., 2010), and similar mechanisms are likely to be involved in the repression of nonendogenous latent viruses. KAP1 restricts HIV1 integration (Allouch et al., 2011), and maintains latency for KSHV (Cai et al., 2013; Gjyshi et al., 2015; King et al., 2015; Sun et al., 2014; Zhang et al., 2014), EBV (Bentz et al., 2015), and HCMV (Rauwel et al., 2015). Phosphorylation of KAP1 decreases its ability to induce heterochromatin and triggers reactivation of HCMV (Rauwel et al., 2015). Sumoylation of KAP1 by LMP1-dependent NF-kB signaling increases the epigenetic silencing of EBV lytic origin (Bentz et al., 2015). The KSHV LANA protein interacts with a SUMO-2 modified KAP1 protein to repress viral reactivation from latency (Cai et al., 2013). The LANA-KAP1 complex was found to associate with the viral immediate early control region upstream of the viral immediate early transcriptional activator (RTA) gene (Sun et al., 2014) and regulate the hypoxia response pathway mediated by the transcriptional regulators HIF1α and RBP-jK (Zhang et al., 2014). Modulation of the KAP1 pathway by the transcription factors NRF2 (Gjyshi et al., 2015), and STAT3 (King et al., 2015) also contribute to KSHV latent to lytic switch, further highlighting the central role of KAP1 in regulating viral latency.
CTCF and Higher-Order Chromatin Organization
The chromatin organizing factor CTCF has been implicated in the control EBV, KSHV, HSV, CMV, and HPV latency (reviewed in Bloom et al., 2010; Pentland and Parish, 2015; Tempera and Lieberman, 2014). For EBV and KSHV, CTCF mediates the recruitment of cohesins and the formation of higher-order DNA conformational structures that have been implicated in regulating the latent to lytic switch, and latency-associated transcription programming. Recent studies with HPV reveal that CTCF regulates the alternative splicing of viral RNA transcripts (Paris et al., 2015), as well as the recruitment of the cohesin subunit SMC1 required for viral genome amplification. CTCF can also bind to the proviral genome of the retrovirus HTLV-1 to regulate RNA splicing, epigenetic boundaries, and the antisense transcription of the viral HBZ gene, necessary for clonal persistence (Satou et al., 2016). Thus, CTCF may facilitate viral chromatin structure, RNA polymerase II programming, and recombination-based genome amplification essential for maintaining latent viral genomes.
DNA Methylation
DNA methylation typically is a mechanism to repress transcription and stabilize viral latency. However, DNA methylation of the EBV genome is paradoxically required for lytic reactivation (Hammerschmidt, 2015). The EBV-encoded ZTA immediate early protein binds preferentially to methylated DNA regulatory elements required for lytic transcription activation (Bhende et al., 2004). Some regions of the EBV latent episome are spared DNA methylation. The EBNA1 promoter, Qp, escapes DNA methylation in all forms of latent infection studied (Bakos et al., 2007). This protected status is imposed by the combined binding of EBNA1 and CTCF (Tempera et al., 2010), leaving this promoter accessible for cell cycle and interferon regulatory control signals during latent infection.
Several viral proteins interact with host DNA methylation machinery. KSHV LANA interacts with DNA methyltransferase 3A (DNMT3A) to facilitate epigenetic silencing of host genes (Shamay et al., 2006). HSV-1 virion proteins VP26 and VP5 interacts with DNMT3A (Rowles et al., 2015). However, shRNA depletion of DNMT3A reduced HSV replication, suggesting that host DNA methylases contribute positively to virus production. Different patterns of viral genome methylation may also explain various gene expression programs for HPV (Doeberitz and Vinokurova, 2009). For example, hypermethylation of HPV E2 binding sites during differentiation can enhance transcription of the viral E6/E7 proteins, which mediate cellular transformation (Vinokurova and von Knebel Doeberitz, 2011).
Epigenetics and Metabolism
Viral latency is found commonly in long-lived, nondividing cells. The metabolism associated with cellular quiescence is likely to be part of the long-term strategy of viral latency. One well studied epigenetic regulator that integrates metabolic information is SIRT1, an NAD-dependent histone deacetylase. SIRT1 has been implicated in the chromatin control of several different viruses, including HIV (Pagans et al., 2005; Pinzone et al., 2013), HBV (Belloni et al., 2009; Li et al., 2015; Ren et al., 2014), HTLV-1 (Tang et al., 2015; Zhang and Wu, 2009), and KSHV (Li et al., 2014). A recent study with KSHV revealed that SIRT1 can interact with the viral genome and viral RTA, to mediate epigenetic silencing. SIRT1 has been shown to deacetylate the HIV transactivator protein, TAT, to reduce its functional activity (Kwon and Ott, 2008). Other NAD-consuming factors, like poly-ADP ribose polymerase (PARP1), have been implicated in control of latency gene expression and replication (Cheong et al., 2015; Ohsaki et al., 2004). How NAD and other cellular metabolites affect epigenetic regulators and viral latency is not yet known.
Noncoding RNAs
Viral noncoding RNAs are common and abundant factors expressed during latency. Numerous functions have been assigned to these noncoding RNAs, including epigenetic control of latent virus and modulation of host processes important for viral latency. HSV1 latency-associated transcript (LAT) is a long noncoding RNA that generates a stable 1.5 kB lariat as a splice product in the neuronal nucleus (reviewed in Kennedy et al., 2015). A major function of LAT is to protect neurons from apoptosis. LAT also provides other functions during latent infection, including maintaining epigenetic silencing on the viral genome. LAT can promote H3K9me3 (Wang et al., 2005) and H3K27me3-dependent heterochromatin on viral lytic cycle genes (Cliffe et al., 2013; Kwiatkowski et al., 2009). On the other hand, LAT can facilitate reactivation from latency (Inman et al., 2001; Perng et al., 2000). How changes in LAT expression or processing correlate with a latent to lytic switch is not fully understood. LAT is also a source of HSV1-encoded miRNAs, several of which have been implicated in regulating host, as well as viral gene targets important for the control of latency.
During all forms of EBV latency, two small noncoding RNAs, termed EBER1 and EBER2, are expressed as RNA polymerase III transcripts. Several functions have been ascribed to EBERS, including the enhancement of tumorigenesis through intrinsic and extrinsic signaling by the innate immune receptor TLR3 (reviewed in Iwakiri, 2014). More recently, EBER2 was shown to have an epigenetic function in controlling latent cycle gene expression (Lee et al., 2015a). EBER2 was found to recruit the transcription factor PAX5 to the viral terminal repeats through a conserved sequence homology between these two different viral loci. Although the interaction with Pax5 is mediated by an unknown factor, the interaction between EBER2 RNA and the nascent transcripts initiating in the terminal repeat were shown to be direct RNA-RNA hybrid complementarity. These finding reveal roles for noncoding and coding RNA-RNA interactions that direct very early events in the selection of transcription factor binding sites during latent infection.
The HCMV noncoding RNA beta 2.7 is expressed in both latent and lytic infected cells (Rossetto et al., 2013a), and has been shown to inhibit apoptosis during lytic infection in myeloid precursor cells, through a mechanism involving the mitochondrial complex I (Reeves et al., 2007). Another HCMV noncoding RNA lnc4.9 interacts with the Polycomb protein complex PRC2 to promote histone H3K27methylation of MIEP (Rossetto et al., 2013a), performing a similar function to HSV1 LAT.
Viral miRNAs
Viral encoded miRNAs have been identified for almost all persistent DNA viruses, including adenoviruses, polyomaviruses, and retroviruses, but the large majority of miRNA are encoded by herpesviruses (Grey, 2015). There is substantial evidence supporting a role of some miRNAs in maintaining viral latency, either directly or indirectly. Most of the targets of viral encoded miRNAs have been shown to be host mRNAs, but there is also evidence for viral encoded miRNAs targeting viral genes to suppress lytic cycle and promote latent infection. For example, HCMV miRNA UL112-1 targets the HCMV IE72 trans-activating gene (Grey et al., 2007; O’Connor et al., 2014), KSHV miR-K12-9* targets the KSHV immediate early transactivator RNA (Bellare and Ganem, 2009), EBV BART20-5p targets EBV immediate-early genes BZLF1 and BRLF1 (Jung et al., 2014), the KSHV K12-7-5p targets KSHV ORF50/RTA (Lin et al., 2011), HSV miRNAs target ICP0 and ICP34.5 during latent infection in neuronal culture (Tang et al., 2009), and latently infected ganglia (Umbach et al., 2008).
In addition, host miRNAs have been identified that regulate viral gene function, such as mir200 family inhibiting HCMV reactivation by targeting viral IE86 gene target that is essential for virus replication (O’Connor et al., 2014). Similarly, a neuronal-specific miR138 targets the HSV1 transcriptional activator ICP0 to suppress lytic gene expression (Pan et al., 2014). Thus, viral and host miRNAs can target viral mRNAs to fine tune viral gene expression and stabilize latent infection.
Reactivation Mechanisms
Reactivation from latency can be triggered by various host-cell intrinsic factors, like terminal differentiation, as well as extrinsic environmental stress, like hypoxia and inflammation (Figure 3). The exit from latency, also referred to as animation, can be distinguished from subsequent steps in the reactivation process (Penkert and Kalejta, 2012). For HCMV, the viral tegument protein pp71 is required prior to immediate early gene activation upon reactivation from latency in CD34+ cells (Penkert and Kalejta, 2010; Qin et al., 2013). However, in certain cell types, pp71 can be sequestered in cytoplasm and degraded by the host protease granzyme M (van Domselaar et al., 2010), thus serving an antiviral function by limiting viral reactivation (Saffert et al., 2010). HSV1 reactivation from sensory neurons requires the de novo synthesis of virion protein VP16 (Thompson et al., 2009) and the relocalization of HCF1 into the nucleus to animate latent viral genomes (Kim et al., 2012; Whitlow and Kristie, 2009). Animation of EBV from latency also requires de novo protein synthesis prior to viral immediate early gene activation (BZLF1 or BRLF1), but it is not clear if this involves viral or cellular proteins (Ye et al., 2007). Gene expression during the early stages of reactivation may be different than that observed during the early stages of primary infection. Upon HSV1 animation from latency, the entire genome undergoes a global derepression with unregulated expression of all temporal classes of viral genes (Du et al., 2011; Kim et al., 2012; Kobayashi et al., 2012). A neuronal-specific Jun-N-terminal kinase (JNK) stress response pathway can rapidly reverse HSV1 epigenetic silencing through the phosphorylation of H3pS10, which can bypass H3K27me3 and H3K9me3 mediated repression by a phosphor-methyl switch mechanism (Cliffe et al., 2015). A second stage of gene expression ensues when VP16 dependent transcription coordinates the temporal regulation of viral lytic genes (Sawtell et al., 2011). However, single cell examination of HSV1 latently infected mouse ganglia shows that lytic gene expression occurs frequently and asynchronously among a population of infected neurons (Ma et al., 2014). This raises some question as to the temporal control of reactivation in a single cell relative to a population average.
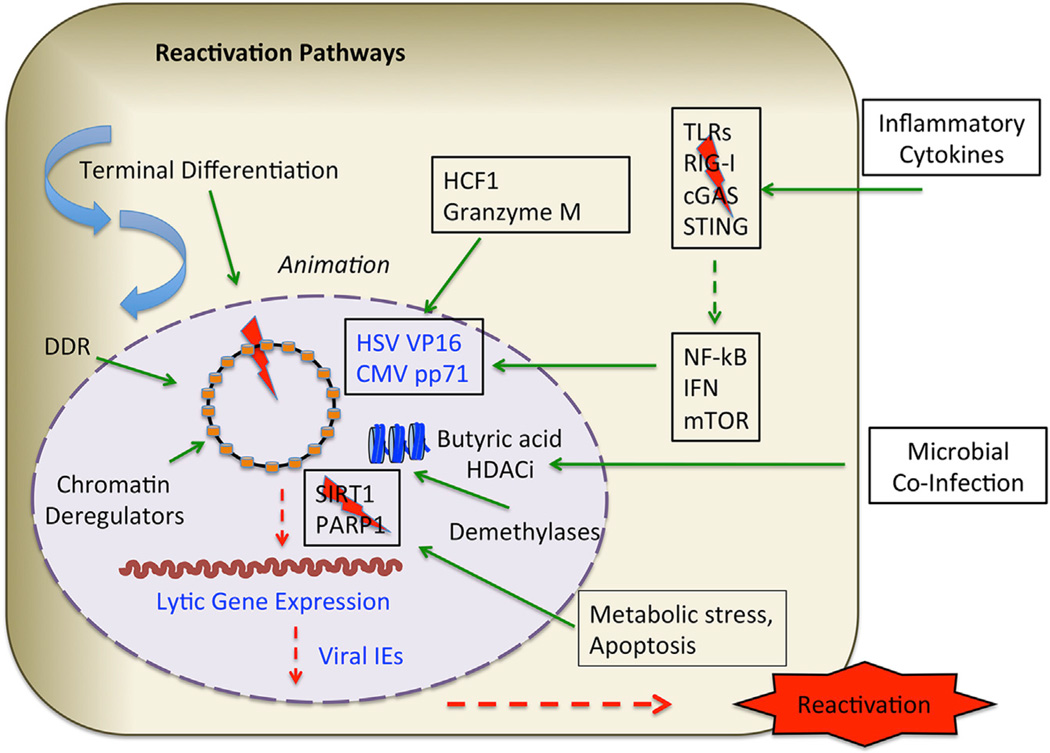
Figure 3. Reactivation Pathways.
Reactivation from latency can be initiated from various extrinsic and intrinsic signals, including host cell terminal differentiation, DNA damage and repair signaling (DDR), and various epigenetic deregulators that perturb viral chromatin. Metabolic stress and apoptosis can alter the activity of chromatin regulatory factors, like SIRT1 and PARP1. Both DNA and histones can be subject to demethylation to reactivate viral lytic cycle gene expression. Host factors, including HCF1 and Granzyme M, can induce expression of viral tegument proteins to animate the early stages of reactivation. Inflammatory cytokines can alter the balance innate immune signaling to promote lytic cycle gene activation. Partial lytic gene expression may occur frequently among populations of latently infected cells (see text for details).
Potential for New Therapies
Several new methods and strategies have been designed to selectively target latent viruses. These include the use of the CRISPR/Cas9 genome editing to selectively delete or mutate latent virus DNA (White et al., 2015). A proof of principle experiment has shown that EBV genomes can be selectively eliminated from latently infected cells in some Burkitt lymphoma cell lines models (Wang and Quake, 2014). Inhibiting viral reactivation from latency has also shown promise. This “locking in latency” approach has been demonstrated for HSV1, where pharmacological inhibitors of the LSD1 histone demethylase prevent shedding and recurrence by promoting the epigenetic suppression of viral genomes (Hill et al., 2014; Liang et al., 2009). The opposite approach of “lytic therapy” (Shirakawa et al., 2013) uses similar epigenetic regulators to release latent virus into a lytic infection. Lytic reactivated virus can be selectively targeted by existing antiviral drugs and as adjuvants for immunological therapies. Another approach has been suggested by studies that reveal CMV latency associated UL138 gene causes the downregulation of the cellular MRP1 drug transporter. Consequently, cells latently infected with HCMV are more sensitive to chemotoxic agents, like vincristine (Weekes et al., 2013). There are also efforts to target viral-specific noncoding RNAs associated with latent infection (Saayman et al., 2015). Since noncoding RNAs, like LAT or EBERs, are known to play key roles in regulating virus latency, this approach could have high therapeutic potential.
Conclusions
This review is far from comprehensive or complete and yet hopefully shows the enormous complexity of viral latency and its regulation at the genetic and epigenetic levels. This information provides great opportunity for the development of innovative and highly selective therapeutic intervention. As viral latency is responsible for life-long pathogenesis and mortality risk, the tasks ahead are in sight, but challenges remain.
Acknowledgments
P.L. is a founder of Vironika, LLC. This review is dedicated to the memory of Greg Pari, whose work and friendship inspired new ways of thinking about unified themes in viral latency.
REFERENCES
- Abraham CG, Kulesza CA. Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models. J. Virol. 2013;87:13193–13205. doi: 10.1128/JVI.02420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allday MJ. EBV finds a polycomb-mediated, epigenetic solution to the problem of oncogenic stress responses triggered by infection. Front. Genet. 2013;4:212. doi: 10.3389/fgene.2013.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe. 2011;9:484–495. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Andrisani OM. Deregulation of epigenetic mechanisms by the hepatitis B virus X protein in hepatocarcinogenesis. Viruses. 2013;5:858–872. doi: 10.3390/v5030858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle JH, Kristie TM. Epigenetic repression of herpes simplex virus infection by the nucleosome remodeler CHD3. MBio. 2014;5:e01027-13. doi: 10.1128/mBio.01027-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakos A, Banati F, Koroknai A, Takacs M, Salamon D, Minarovits-Kormuta S, Schwarzmann F, Wolf H, Niller HH, Minarovits J. High-resolution analysis of CpG methylation and in vivo protein-DNA interactions at the alternative Epstein-Barr virus latency promoters Qp and Cp in the nasopharyngeal carcinoma cell line C666-1. Virus Genes. 2007;35:195–202. doi: 10.1007/s11262-007-0095-y. [DOI] [PubMed] [Google Scholar]
- Ballestas ME, Kaye KM. The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Future Microbiol. 2011;6:1399–1413. doi: 10.2217/fmb.11.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzile JP, Stark TJ, Yeo GW, Spector DH. Human cytomegalovirus infection of human embryonic stem cell-derived primitive neural stem cells is restricted at several steps but leads to the persistence of viral DNA. J. Virol. 2014;88:4021–4039. doi: 10.1128/JVI.03492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz GL, Moss CR, 2nd, Whitehurst CB, Moody CA, Pagano JS. LMP1-Induced Sumoylation Influences the Maintenance of Epstein-Barr Virus Latency through KAP1. J. Virol. 2015;89:7465–7477. doi: 10.1128/JVI.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 2004;36:1099–1104. doi: 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]
- Bloom DC, Giordani NV, Kwiatkowski DL. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta. 2010;1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J. Mol. Biol. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 2013;94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- Cai Q, Cai S, Zhu C, Verma SC, Choi JY, Robertson ES. A unique SUMO-2-interacting motif within LANA is essential for KSHV latency. PLoS Pathog. 2013;9:e1003750. doi: 10.1371/journal.ppat.1003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarena V, Kobayashi M, Kim JY, Roehm P, Perez R, Gardner J, Wilson AC, Mohr I, Chao MV. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe. 2010;8:320–330. doi: 10.1016/j.chom.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong WC, Park JH, Kang HR, Song MJ. Downregulation of poly(adp-ribose) polymerase 1 by a viral processivity factor facilitates lytic replication of gammaherpesvirus. J. Virol. 2015;89:9676–9682. doi: 10.1128/JVI.00559-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe AR, Coen DM, Knipe DM. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. MBio. 2013;4:e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe AR, Arbuckle JH, Vogel JL, Geden MJ, Rothbart SB, Cusack CL, Strahl BD, Kristie TM, Deshmukh M. Neuronal stress pathway mediating a histone methyl/phospho switch is required for herpes simplex virus reactivation. Cell Host Microbe. 2015;18:649–658. doi: 10.1016/j.chom.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- Dictor M, Rambech E, Way D, Witte M, Bendsöe N. Human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) DNA in Kaposi’s sarcoma lesions, AIDS Kaposi’s sarcoma cell lines, endothelial Kaposi’s sarcoma simulators, and the skin of immunosuppressed patients. Am. J. Pathol. 1996;148:2009–2016. [PMC free article] [PubMed] [Google Scholar]
- Doeberitz Mv, Vinokurova S. Host factors in HPV-related carcinogenesis: cellular mechanisms controlling HPV infections. Arch. Med. Res. 2009;40:435–442. doi: 10.1016/j.arcmed.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Du T, Zhou G, Khan S, Gu H, Roizman B. Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc. Natl. Acad. Sci. USA. 2010;107:15904–15909. doi: 10.1073/pnas.1010741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zhou G, Roizman B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc. Natl. Acad. Sci. USA. 2011;108:18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducroux A, Benhenda S, Rivière L, Semmes OJ, Benkirane M, Neuveut C. The Tudor domain protein Spindlin1 is involved in intrinsic antiviral defense against incoming hepatitis B Virus and herpes simplex virus type 1. PLoS Pathog. 2014;10:e1004343. doi: 10.1371/journal.ppat.1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RH, Sitki-Green D, Moore DT, Raab-Traub N. Potential selection of LMP1 variants in nasopharyngeal carcinoma. J. Virol. 2004;78:868–881. doi: 10.1128/JVI.78.2.868-881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Farrell PJ. Epstein-Barr Virus Strain Variation. Curr. Top. Microbiol. Immunol. 2015;390:45–69. doi: 10.1007/978-3-319-22822-8_4. [DOI] [PubMed] [Google Scholar]
- Feng FT, Cui Q, Liu WS, Guo YM, Feng QS, Chen LZ, Xu M, Luo B, Li DJ, Hu LF, et al. A single nucleotide polymorphism in the Epstein-Barr virus genome is strongly associated with a high risk of nasopharyngeal carcinoma. Chinese J. Cancer. 2015;34:61. doi: 10.1186/s40880-015-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish KN, Stenglein SG, Ibanez C, Nelson JA. Cytomegalovirus persistence in macrophages and endothelial cells. Scand. J. Infect. Dis. Suppl. 1995;99:34–40. [PubMed] [Google Scholar]
- Flint SJ, Racaniello VR, Rall GF, Skalka AM, Enquist LW. Principles of Virology. 4th. Washington, DC: ASM Press; 2015. [Google Scholar]
- Frappier L. Ebna1. Curr. Top. Microbiol. Immunol. 2015;391:3–34. doi: 10.1007/978-3-319-22834-1_1. [DOI] [PubMed] [Google Scholar]
- Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 2011;85:9078–9089. doi: 10.1128/JVI.00836-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennart I, Coupeau D, Pejaković S, Laurent S, Rasschaert D, Muylkens B. Marek’s disease: genetic regulation of gallid herpesvirus 2 infection and latency. Vet. J. 2015;205:339–348. doi: 10.1016/j.tvjl.2015.04.038. [DOI] [PubMed] [Google Scholar]
- Gjyshi O, Roy A, Dutta S, Veettil MV, Dutta D, Chandran B. Activated Nrf2 interacts with Kaposi’s sarcoma-associated herpesvirus latency protein lana-1 and host protein KAP1 to mediate global lytic gene repression. J. Virol. 2015;89:7874–7892. doi: 10.1128/JVI.00895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A, Fester N, Engel AT, Kaufer BB. Role of the short telomeric repeat region in Marek’s disease virus replication, genomic integration, and lymphomagenesis. J. Virol. 2014;88:14138–14147. doi: 10.1128/JVI.02437-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F. Role of microRNAs in herpesvirus latency and persistence. J. Gen. Virol. 2015;96:739–751. doi: 10.1099/vir.0.070862-0. [DOI] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullo C, Low WK, Teoh G. Association of Epstein-Barr virus with nasopharyngeal carcinoma and current status of development of cancer-derived cell lines. Ann. Acad. Med. Singapore. 2008;37:769–777. [PubMed] [Google Scholar]
- Günther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadinoto V, Shapiro M, Sun CC, Thorley-Lawson DA. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 2009;5:e1000496. doi: 10.1371/journal.ppat.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W. The epigenetic life cycle of Epstein-Barr virus. Curr. Top. Microbiol. Immunol. 2015;390:103–117. doi: 10.1007/978-3-319-22822-8_6. [DOI] [PubMed] [Google Scholar]
- Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, et al. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci. Transl. Med. 2014;6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland PL, McDade SS, McCloskey R, Dickson GJ, Arthur K, McCance DJ, Patel D. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J. Virol. 2011;85:10999–11006. doi: 10.1128/JVI.00160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman M, Perng GC, Henderson G, Ghiasi H, Nesburn AB, Wechsler SL, Jones C. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 2001;75:3636–3646. doi: 10.1128/JVI.75.8.3636-3646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D. Epstein-Barr virus-encoded RNAs: key molecules in viral pathogenesis. Cancers (Basel) 2014;6:1615–1630. doi: 10.3390/cancers6031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Corey L. Current concepts for genital herpes simplex virus infection: diagnostics and pathogenesis of genital tract shedding. Clin. Microbiol. Rev. 2016;29:149–161. doi: 10.1128/CMR.00043-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YJ, Choi H, Kim H, Lee SK. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J. Virol. 2014;88:9027–9037. doi: 10.1128/JVI.00721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufer BB, Jarosinski KW, Osterrieder N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 2011;208:605–615. doi: 10.1084/jem.20101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly GL, Long HM, Stylianou J, Thomas WA, Leese A, Bell AI, Bornkamm GW, Mautner J, Rickinson AB, Rowe M. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 2009;5:e1000341. doi: 10.1371/journal.ppat.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PG, Rovnak J, Badani H, Cohrs RJ. A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation. J. Gen. Virol. 2015;96:1581–1602. doi: 10.1099/vir.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Mandarino A, Chao MV, Mohr I, Wilson AC. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog. 2012;8:e1002540. doi: 10.1371/journal.ppat.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CA, Li X, Barbachano-Guerrero A, Bhaduri-McIntosh S. STAT3 regulates lytic activation of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2015;89:11347–11355. doi: 10.1128/JVI.02008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ. Pathogenetic mechansims of recurrent herpes simplex virus infections. Arch. Virol. 1976;51:1–13. doi: 10.1007/BF01317829. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Kim JY, Camarena V, Roehm PC, Chao MV, Wilson AC, Mohr I. A primary neuron culture system for the study of herpes simplex virus latency and reactivation. J. Vis. Exp. 2012 doi: 10.3791/3823. http://dx.doi.org/10.3791/3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie TM. Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology. 2015;479–480:555–561. doi: 10.1016/j.virol.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DL, Thompson HW, Bloom DC. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 2009;83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Ott M. The ups and downs of SIRT1. Trends Biochem. Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015a;160:607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Albright ER, Lee JH, Jacobs D, Kalejta RF. Cellular defense against latent colonization foiled by human cytomegalovirus UL138 protein. Sci. Adv. 2015b;1:e1501164. doi: 10.1126/sciadv.1501164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, He M, Zhou F, Ye F, Gao SJ. Activation of Kaposi’s sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: identification of sirtuin 1 as a regulator of the KSHV life cycle. J. Virol. 2014;88:6355–6367. doi: 10.1128/JVI.00219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WY, Ren JH, Tao NN, Ran LK, Chen X, Zhou HZ, Liu B, Li XS, Huang AL, Chen J. The SIRT1 inhibitor, nicotinamide, inhibits hepatitis B virus replication in vitro and in vivo. Arch. Virol. 2015;161:621–630. doi: 10.1007/s00705-015-2712-8. [DOI] [PubMed] [Google Scholar]
- Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. miR-K12-7-5p encoded by Kaposi’s sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS ONE. 2011;6:e16224. doi: 10.1371/journal.pone.0016224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JZ, Russell TA, Spelman T, Carbone FR, Tscharke DC. Lytic gene expression is frequent in HSV-1 latent infection and correlates with the engagement of a cell-intrinsic transcriptional response. PLoS Pathog. 2014;10:e1004237. doi: 10.1371/journal.ppat.1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Kobayashi-Ishihara M, Fujikawa D, Ishida T, Watanabe T, Yamagishi M. Epigenetic heterogeneity in HIV-1 latency establishment. Sci. Rep. 2015;5:7701. doi: 10.1038/srep07701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG. Nuclear domain 10, the site of DNA virus transcription and replication. Bio Essays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- McBride AA. The papillomavirus E2 proteins. Virology. 2013;445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Shenk T. Human cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 2008;325:1–19. doi: 10.1007/978-3-540-77349-8_1. [DOI] [PubMed] [Google Scholar]
- O’Connor CM, Vanicek J, Murphy EA. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J. Virol. 2014;88:5524–5532. doi: 10.1128/JVI.00481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki E, Ueda K, Sakakibara S, Do E, Yada K, Yamanishi K. Poly(ADP-ribose) polymerase 1 binds to Kaposi’s sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 2004;78:9936–9946. doi: 10.1128/JVI.78.18.9936-9946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J. Virol. 2015;89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe. 2014;15:446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris C, Pentland I, Groves I, Roberts DC, Powis SJ, Coleman N, Roberts S, Parish JL. CCCTC-binding factor recruitment to the early region of the human papillomavirus 18 genome regulates viral oncogene expression. J. Virol. 2015;89:4770–4785. doi: 10.1128/JVI.00097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert RR, Kalejta RF. Nuclear localization of tegument-delivered pp71 in human cytomegalovirus-infected cells is facilitated by one or more factors present in terminally differentiated fibroblasts. J. Virol. 2010;84:9853–9863. doi: 10.1128/JVI.00500-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert RR, Kalejta RF. Tale of a tegument transactivator: the past, present and future of human CMV pp71. Future Virol. 2012;7:855–869. doi: 10.2217/fvl.12.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentland I, Parish JL. Targeting CTCF to control virus gene expression: a common theme amongst diverse DNA viruses. Viruses. 2015;7:3574–3585. doi: 10.3390/v7072791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- Pinkevych M, Cromer D, Tolstrup M, Grimm AJ, Cooper DA, Lewin SR, Søgaard OS, Rasmussen TA, Kent SJ, Kelleher AD, Davenport MP. HIV reactivation from latency after treatment interruption occurs on average every 5–8 days—implications for HIV remission. PLoS Pathog. 2015;11:e1005000. doi: 10.1371/journal.ppat.1005000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzone MR, Cacopardo B, Condorelli F, Di Rosa M, Nunnari G. Sirtuin-1 and HIV-1: an overview. Curr. Drug Targets. 2013;14:648–652. doi: 10.2174/1389450111314060005. [DOI] [PubMed] [Google Scholar]
- Poole E, Sinclair J. Sleepless latency of human cytomegalovirus. Med. Microbiol. Immunol. (Berl.) 2015;204:421–429. doi: 10.1007/s00430-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Penkert RR, Kalejta RF. Heterologous viral promoters incorporated into the human cytomegalovirus genome are silenced during experimental latency. J. Virol. 2013;87:9886–9894. doi: 10.1128/JVI.01726-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. eLife. 2015;4 doi: 10.7554/eLife.06068. http://dx.doi.org/10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- Ren JH, Tao Y, Zhang ZZ, Chen WX, Cai XF, Chen K, Ko BC, Song CL, Ran LK, Li WY, et al. Sirtuin 1 regulates hepatitis B virus transcription and replication by targeting transcription factor AP-1. J. Virol. 2014;88:2442–2451. doi: 10.1128/JVI.02861-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J. Hepatol. 2015;63:1093–1102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Robinson CM, Hunt HD, Cheng HH, Delany ME. Chromosomal integration of an avian oncogenic herpesvirus reveals telomeric preferences and evidence for lymphoma clonality. Herpesviridae. 2010;1:5. doi: 10.1186/2042-4280-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Zhou G, Du T. Checkpoints in productive and latent infections with herpes simplex virus 1: conceptualization of the issues. J. Neurovirol. 2011;17:512–517. doi: 10.1007/s13365-011-0058-x. [DOI] [PubMed] [Google Scholar]
- Rossetto CC, Pari GS. PAN’s Labyrinth: Molecular biology of Kaposi’s sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses. 2014;6:4212–4226. doi: 10.3390/v6114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013a;9:e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013b;87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowles DL, Tsai YC, Greco TM, Lin AE, Li M, Yeh J, Cristea IM. DNA methyltransferase DNMT3A associates with viral proteins and impacts HSV-1 infection. Proteomics. 2015;15:1968–1982. doi: 10.1002/pmic.201500035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Roberts TC, Morris KV, Weinberg MS. HIV Latency and the noncoding RNA therapeutic landscape. Adv. Exp. Med. Biol. 2015;848:169–189. doi: 10.1007/978-1-4939-2432-5_9. [DOI] [PubMed] [Google Scholar]
- Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 2006;80:3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert RT, Kalejta RF. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 2007;81:9109–9120. doi: 10.1128/JVI.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert RT, Penkert RR, Kalejta RF. Cellular and viral control over the initial events of human cytomegalovirus experimental latency in CD34+ cells. J. Virol. 2010;84:5594–5604. doi: 10.1128/JVI.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Miyazato P, Ishihara K, Yaguchi H, Melamed A, Miura M, Fukuda A, Nosaka K, Watanabe T, Rowan AG, et al. The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1423199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell NM, Triezenberg SJ, Thompson RL. VP16 serine 375 is a critical determinant of herpes simplex virus exit from latency in vivo. J. Neurovirol. 2011;17:546–551. doi: 10.1007/s13365-011-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamay M, Krithivas A, Zhang J, Hayward SD. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi’s sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. USA. 2006;103:14554–14559. doi: 10.1073/pnas.0604469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol. 2013;21:277–285. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck SH, Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Liang D, Gao Y, Lan K. Kaposi’s sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency. J. Virol. 2014;88:7331–7344. doi: 10.1128/JVI.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Patel A, Krause PR. Novel less-abundant viral micro-RNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HM, Gao WW, Chan CP, Cheng Y, Deng JJ, Yuen KS, Iha H, Jin DY. SIRT1 suppresses human t-cell leukemia virus type 1 transcription. J. Virol. 2015;89:8623–8631. doi: 10.1128/JVI.01229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant-Elorza M, Rossetto CC, Pari GS. Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe. 2014;16:43–54. doi: 10.1016/j.chom.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Taylor-Wiedeman J, Sissons P, Sinclair J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempera I, Lieberman PM. Epigenetic regulation of EBV persistence and oncogenesis. Semin. Cancer Biol. 2014;26:22–29. doi: 10.1016/j.semcancer.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 2010;6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RL, Preston CM, Sawtell NM. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog. 2009;5:e1000352. doi: 10.1371/journal.ppat.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA. EBV Persistence–Introducing the Virus. Curr. Top. Microbiol. Immunol. 2015;390:151–209. doi: 10.1007/978-3-319-22822-8_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010;6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Brulois K, Lee HR, Izumiya Y, Tepper C, Kung HJ, Jung JU. Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection. PLoS Pathog. 2013;9:e1003813. doi: 10.1371/journal.ppat.1003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc. Natl. Acad. Sci. USA. 2015;112:E5715–E5724. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MH, Raykova A, Klinke O, Bernhardt K, Gärtner K, Leung CS, Geletneky K, Sertel S, Münz C, Feederle R, Delecluse HJ. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell Rep. 2013;5:458–470. doi: 10.1016/j.celrep.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Tsai K, Chan L, Gibeault R, Conn K, Dheekollu J, Domsic J, Marmorstein R, Schang LM, Lieberman PM. Viral reprogramming of the Daxx histone H3.3 chaperone during early Epstein-Barr virus infection. J. Virol. 2014;88:14350–14363. doi: 10.1128/JVI.01895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Messick TE, Lieberman PM. Disruption of host antiviral resistances by gammaherpesvirus tegument proteins with homology to the FGARAT purine biosynthesis enzyme. Curr. Opin. Virol. 2015;14:30–40. doi: 10.1016/j.coviro.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao SW, Tsang CM, To KF, Lo KW. The role of Epstein-Barr virus in epithelial malignancies. J. Pathol. 2015;235:323–333. doi: 10.1002/path.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. KSHV LANA–the master regulator of KSHV latency. Viruses. 2014;6:4961–4998. doi: 10.3390/v6124961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Domselaar R, Philippen LE, Quadir R, Wiertz EJ, Kummer JA, Bovenschen N. Noncytotoxic inhibition of cytomegalovirus replication through NK cell protease granzyme M-mediated cleavage of viral phosphoprotein 71. J. Immunol. 2010;185:7605–7613. doi: 10.4049/jimmunol.1001503. [DOI] [PubMed] [Google Scholar]
- Vinokurova S, von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS ONE. 2011;6:e24451. doi: 10.1371/journal.pone.0024451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc. Natl. Acad. Sci. USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA. 2005;102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson Z, Dhummakupt A, Messer H, Phelan D, Bloom D. Role of polycomb proteins in regulating HSV-1 latency. Viruses. 2013;5:1740–1757. doi: 10.3390/v5071740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes MP, Tan SY, Poole E, Talbot S, Antrobus R, Smith DL, Montag C, Gygi SP, Sinclair JH, Lehner PJ. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science. 2013;340:199–202. doi: 10.1126/science.1235047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RE, Rämer PC, Naresh KN, Meixlsperger S, Pinaud L, Rooney C, Savoldo B, Coutinho R, Bödör C, Gribben J, et al. EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J. Clin. Invest. 2012;122:1487–1502. doi: 10.1172/JCI58092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Hu W, Khalili K. The CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discov. Med. 2015;19:255–262. [PMC free article] [PubMed] [Google Scholar]
- Whitlow Z, Kristie TM. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J. Virol. 2009;83:9591–9595. doi: 10.1128/JVI.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CL, Smith RL. HSV latency in vitro : in situ hybridization methods. Methods Mol. Med. 1998;10:317–326. doi: 10.1385/0-89603-347-3:317. [DOI] [PubMed] [Google Scholar]
- Ye J, Gradoville L, Daigle D, Miller G. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi’s sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J. Virol. 2007;81:9279–9291. doi: 10.1128/JVI.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Wu MR. SIRT1 regulates Tat-induced HIV-1 trans-activation through activating AMP-activated protein kinase. Virus Res. 2009;146:51–57. doi: 10.1016/j.virusres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhu C, Guo Y, Wei F, Lu J, Qin J, Banerjee S, Wang J, Shang H, Verma SC, et al. Inhibition of KAP1 enhances hypoxia-induced Kaposi’s sarcoma-associated herpesvirus reactivation through RBP-Jκ. J. Virol. 2014;88:6873–6884. doi: 10.1128/JVI.00283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]