Fig. 4.
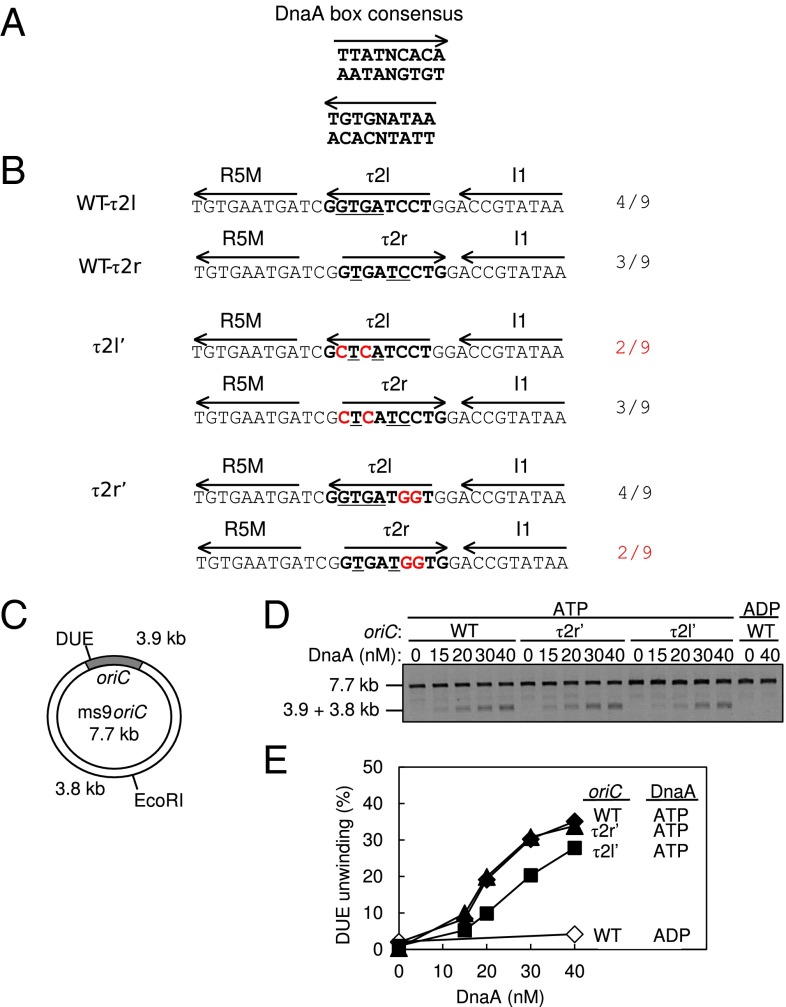
DUE activity of oriC bearing mutant τ2 sequences. (A) DnaA box consensus. The 9-mer consensus sequence with both orientations (arrow) are shown. (B) WT and mutant sequences including the R5M and I1 boxes. The Upper two lines show the WT sequence and each definition of the τ2 box (boldface) with the orientation (arrow). Identical bases to the 9-mer consensus are indicated by underlining; the number is also shown. The Middle two lines show the τ2l′ mutant sequence. The Bottom two lines show the τ2r′ mutant sequence. Each definition of the τ2 box is also shown. The mutations are highlighted by red letters. (C) Structure of an oriC plasmid M13oriCMS9 (ms9oriC). The positions of oriC, DUE, and the EcoRI site are indicated. (D and E) DUE assay. The given amounts of ATP–DnaA or ADP–DnaA were incubated with M13oriCMS9 or its derivative bearing τ2l′ or τ2r′ mutation, followed by analysis using ssDNA-specific nuclease P1, EcoRI, and agarose gel electrophoresis (D). Band intensities of the gel image were quantified and the relative amounts of the 3.8- and 3.9-kb bands were plotted as DUE (%) (E).