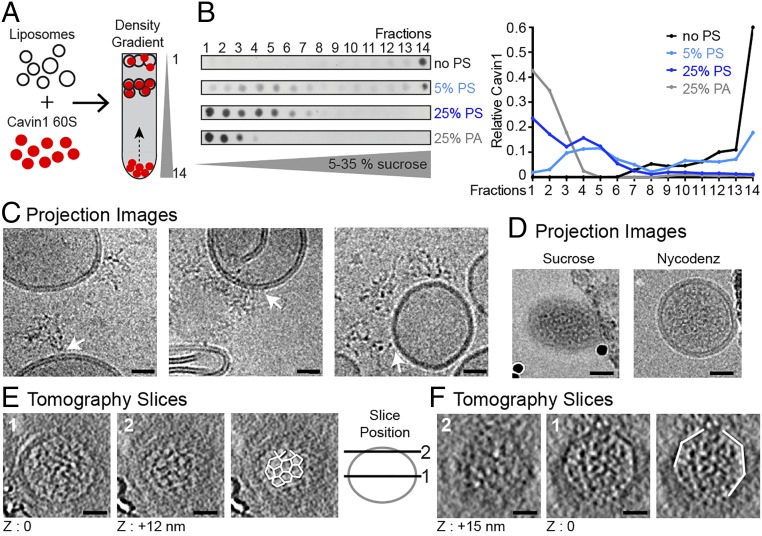
Fig. 2.
The Cavin1 complex associates with liposomes and exhibits locally ordered polygonal architecture. (A) Schematic of Cavin1–liposome incubation and subsequent proteoliposome flotation by density gradient centrifugation. (B) Cavin1 60S complexes were incubated with liposomes containing 5 or 25% PS or 25% PA (in the lipid mix: 25% DOPS, 15% DOPC, 15% DOPE, 45% cholesterol) and floated in 5–35% sucrose density gradients. Gradient fractions were analyzed by dot blot using an anti-HA antibody. The graph shows the quantification of Cavin1 distribution across fractions. (C–F) Cavin1 60S complexes incubated with 25% PS liposomes. (C) CryoEM projection images of liposomes incubated with Cavin1, before flotation. 60S complexes show connections to the liposome bilayer (white arrows). Images were taken at different defocus values. (D–F) CryoEM/ET analysis of proteoliposomes from fractions 4 and 5 of the gradients. (D) 2D projection images with sucrose or Nycodenz as the density medium. Liposomes appear naked on the outside of the membrane. The micrographs of sucrose gradients were recorded on a CCD camera at a higher defocus (−5 to −6 µm) than the micrographs of Nycodenz gradient samples acquired on a K2 summit direct detector (−2 to −4 µm). (E and F) CryoET of two exemplary proteoliposomes. For each, a central (1) and tangential (2) slice through the tomogram is shown. Slice position is depicted in the adjacent scheme. Cavin1 appears filamentous inside the liposome and adopts a polyhedral net-like structure in contact with the membrane (outlined in white in E). Angular shapes of proteoliposomes were visible (white outlines in F). Images were recorded at 200 or 300 keV with a calibrated pixel size of 5.9 Å or 4.9 Å at defoci of −3.5 to −5 µm. (Scale bars: 20 nm.) The thickness of the tomographic slice in E and F is 12 Å. Full tomographic volumes corresponding to E and F are shown in Movies S1 and S2.