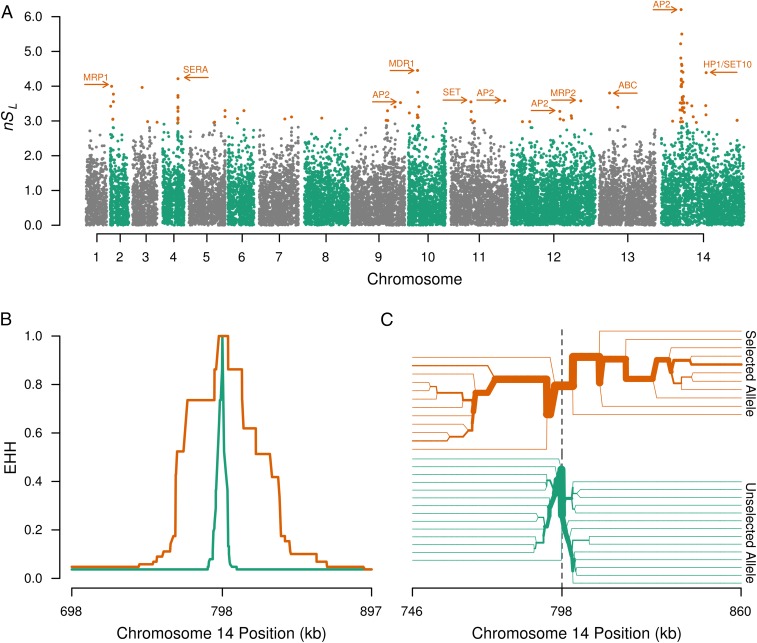
Fig. 5.
Evidence for strong selective sweeps in Cambodian P. vivax. (A) A Manhattan plot of normalized nSL values. Each point corresponds to an SNP, and the top 0.5% values (under strong directional selection) are rendered in orange. Polymorphisms without evidence of strong directional selection are rendered in either gray or green, according to chromosome. This view suggests that several genomic regions are under positive selection, including areas near transcription factors (AP2 domain-containing), chromatin regulators (SET10 and HP1), antigens under known positive selection (SERA4 and 5), and drug-resistance genes (MDR1 and MRP1). (B and C) The extent of EHH in the strongest sweep, within chromosome 14 in A. (B) EHH decay for the haplotypes around selected (orange) and unselected (green) alleles. (C) A haplotype bifurcation diagram is shown centered on the focal variant. Line thickness represents the number of identical haplotypes flanking the selected (orange) or unselected (green) alleles. Linkage breaks down with increasing distance from the focal variant. B and C provide evidence that the strong sweep on chromosome 14 extends ∼50 kb in each direction.