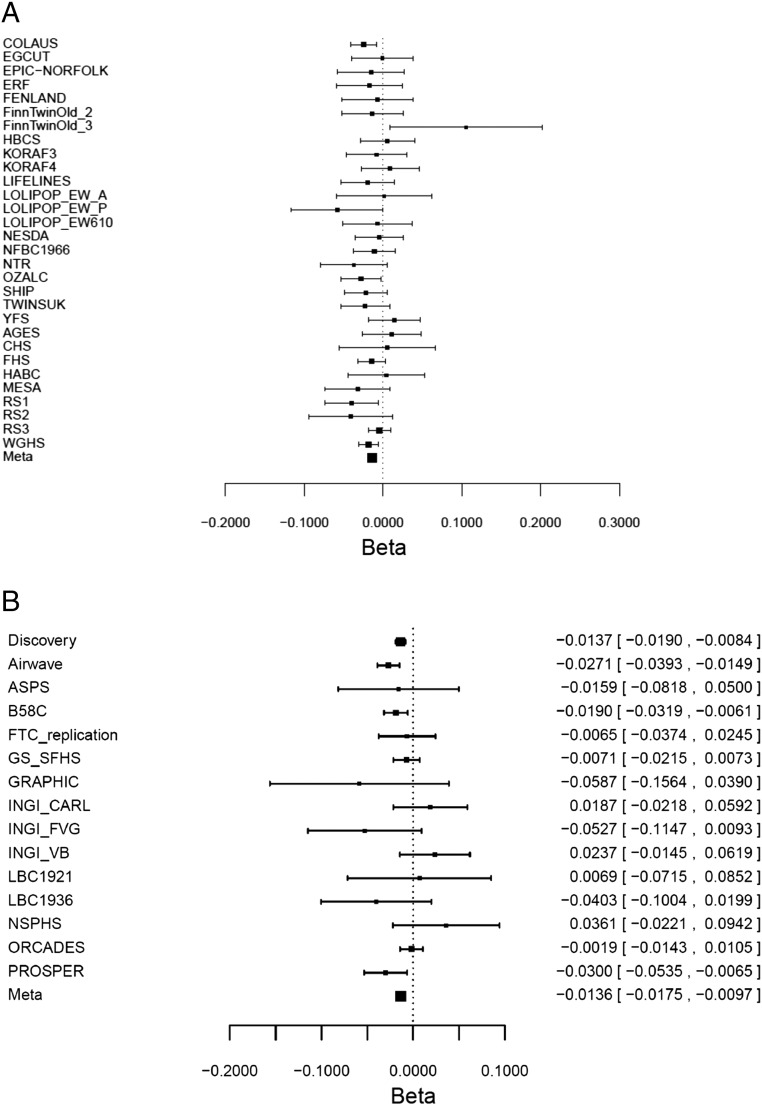
Fig. S1.
Forest plot for the association of rs11940694 in KLB with log grams per day alcohol in the discovery GWAS and replication cohorts. (A) rs11940694 in KLB in discovery GWAS cohorts. Discovery GWAS cohorts: the Alcohol Genome-Wide Association (AlcGen) Consortium: the Cohorte Lausannoise study (COLAUS), the Estonian Biobank Cohort (EGCUT), the European Prospective Investigation of Cancer–Norfolk study (EPIC-NORFOLK), the Erasmus Rucphen Family study (ERF), the Fenland study (FENLAND), the Older Finnish Twin Cohort_2 (FinnTwinOld_2), the Helsinki Birth Cohort Study (HBCS), the population-based Cooperative Health Research in the Region of Augsburg F3 Study (KORAF3), the population-based Cooperative Health Research in the Region of Augsburg F4 Study (KORAF4), the LifeLines Cohort Study & Biobank (LIFELINES), the London Life Sciences Prospective Population Study (LOLIPOP_EW_A, LOLIPOP_EW_P, and LOLIPOP_EW610), the Older Finnish Twin Cohort_3 (FinnTwinOld_3), the Netherlands Study of Depression and Anxiety (NESDA), the Northern Finland Birth Cohort 1966 (NFBC1996), the Netherlands Twin Register cohort (NTR), the Australian twin-family study of alcohol use disorder (OZALC), the Study of Health in Pomerania (SHIP), the TwinsUK study (TWINSUK), and the Cardiovascular Risk in Young Finns Study (YFS); and the Cohorts for Heart and Aging Research in Genomic Epidemiology Plus (CHARGE+) Consortium: the Age, Gene/Environment Susceptibility–Reykjavik (AGES-Reykjavik) study, the Cardiovascular Health Study (CHS), the Framingham Heart Study (FHS), the Health, Aging, and Body Composition (HABC), the Multi-Ethnic Study of Atherosclerosis (MESA), the Rotterdam Study (RS1, RS2, and RS3), and the Women's Genome Health Study (WGHS). In rs11940694, the coded allele was A, and the noncoded allele was G. The allele frequency for A was ∼0.42 in the entire sample. The beta/SE estimates were for A allele. (B) rs11940694 in KLB in discovery and replication cohorts. The coded allele was A, and the noncoded allele was G. The beta/SE estimates were for A allele.