Abstract
Malaria is a life-threatening disease caused by Plasmodium falciparum parasites that is transmitted through the bites of infected anopheline mosquitoes. P. falciparum dispersal from Africa, as a result of human migration, required adaptation of the parasite to several different indigenous anopheline species. The mosquito immune system can greatly limit infection and P. falciparum evolved a strategy to evade these responses that is mediated by the Pfs47 gene. Pfs47 is a polymorphic gene with signatures of diversifying selection and a strong geographic genetic structure at a continental level. Here, we investigated the role of single four amino acid differences between the Pfs47 gene from African (GB4 and NF54) and a New World (7G8) strains that differ drastically in their ability to evade the immune system of A. gambiae L35 refractory mosquitoes. Wild type NF54 and GB4 parasites can survive in this mosquito strain, while 7G8 parasites are eliminated. Our studies indicate that replacement in any of these four single amino acids in Pfs47 from the NF54 strain by those present in 7G8, completely disrupts the ability of NF54 parasites to hide from the mosquito immune system. One of these amino acid replacements had the opposite effect on A. albimanus mosquitoes, and enhanced infection. We conclude that malaria transmission involves a complex interplay between the genetic background of the parasite and the mosquito and that Pfs47 can be critical in this interaction as it mediates Plasmodium immune evasion through molecular interactions that need to be precise in some parasite/vector combinations.
Introduction
Plasmodium falciparum causes the most virulent form of malaria in humans and is transmitted by anopheline mosquitoes. In 2015, there were 214 million new cases and an estimated 438,000 malaria deaths worldwide [1]. P. falciparum malaria appears to have originated in Africa from a single horizontal transfer from gorillas to humans [2] and became a global disease as infected humans migrated out of Africa. This remarkable journey led to the adaptation of the parasite to more than 70 different anopheline vector species [3]. In some cases, parasites encountered mosquito species that were evolutionarily distant from African vectors. For example, anophelines of the subgenus Nyssorhynchus (malaria vectors in Central and South America, such as Anopheles albimanus) diverged from the subgenus Cellia (malaria vectors in Africa, India, and South Asia) about 100 million years ago (MYA) [4].
P. falciparum isolates from different geographic origins can exhibit dramatic differences in infectivity to the same mosquito vector, suggesting that natural selection took place as parasites adapted to different mosquito vector species. Some P. falciparum lines of African origin (GB4 and NF54) can infect an Anopheles gambiae L35 strain (R strain) that was selected to be highly refractory to infection with Plasmodium cynomolgi (monkey malaria), while a line from Brazil (7G8) is eliminated and encapsulated [5, 6]. Conversely, Anopheles albimanus (a New World vector) is more susceptible to infection with 7G8 (Brazilian) than with African P. falciparum lines [7]. Recent studies indicate that the mosquito complement-like system is a major determinant of vector-parasite compatibility. The thioester containing protein-1 (TEP1) is a key component of the A. gambiae complement-like system that binds to the surface of Plasmodium ookinetes, as they come in contact with the mosquito hemolymph, activating the formation of a complex that kills the parasite [8]. In A. gambiae R strain, damaged parasites are covered with melanin [5], an insoluble black pigment, through a TEP1-dependent mechanism, while in other mosquito strains parasites can be lyzed by the complement system. TEP1 is stabilized in the mosquito hemolymph by forming a complex with two leucine-rich repeat proteins, LRIM1 and APL1C [9, 10]. Silencing either LRIM1 or APL1C results in premature, systemic activation and deposition of TEP1 or the active form (TEP1-cut) is depleted in the hemolymph [9, 10]. Disruption of the mosquito complement system in A. gambiae R strain mosquitoes, by silencing either TEP1 or LRIM1, reverts the refractory phenotype and allows P. falciparum 7G8 parasites to survive, while it has no significant effect on the prevalence or intensity of infection with NF54 and GB4 parasites [6]. This indicates that 7G8 parasites are detected and eliminated by the mosquito immune system, while most NF54 and GB4 parasites are able to evade the system and survive.
A combination of genetic linkage mapping in a cross between P. falciparum GB4 (an African strain that survives in A. gambiae R strain) and a P. falciparum 7G8 (Brazilian strain that is melanized), followed by linkage group selection and functional genomics analysis was used to identify Pfs47 as the gene that allows African (GB4) parasites to become “invisible” to the mosquito immune system [11]. Genetic disruption of Pfs47 in P. falciparum NF54 resulted in almost complete elimination of the parasite in A. gambiae that could be reverted by silencing TEP1 expression [11]. Global analysis of 364 different P. falciparum isolates identified 42 different Pfs47 haplotypes with strong geographic structure. Laboratory infections showed that P. falciparum isolates from Africa, Asia, or the Americas have low compatibility to malaria vectors from a different continent that can be overcomed by disrupting the mosquito immune system [7]. Replacement of the Pfs47 gene in NF54 (African) parasites with a Pfs47 haplotype common in Asia or the New World made the African parasite compatible with an Asian (A. dirus) or American (A. albimanus) vector respectively, by making the parasite “undetectable” by the mosquito immune system in these vectors [7]. This indicates that Pfs47 plays a key role in Plasmodium-mosquito compatibility and malaria transmission worldwide. Remarkably, there are only four amino acid differences between the Pfs47 genes in the GB4 and 7G8 lines that determine the survival of the GB4 in A. gambiae R strain. These amino acid differences (T236I, S242L, V247A and I248L) are localized in the second domain (D2) of Pfs47, between the two cysteines in that domain, that are 30 amino acids (aa) apart. In this manuscript, we analyzed the molecular basis of compatibility of Pfs47 and A. gambiae by generating NF54 transformed parasites in which each of the four amino acid differences in Pfs47 between the GB4 and 7G8 strains was introduced by replacing the residues in African NF54 parasites with those of the 7G8 line from Brazil, changing one amino acid at a time. Stable parasites that only differ by a single amino acid with the same genetic background were generated. We hypothesized that at least one of these single amino acid changes would reduce survival of NF54 parasites in A. gambiae R strain mosquitoes. The effect of these mutations on A. albimanus infection was also analyzed.
Results
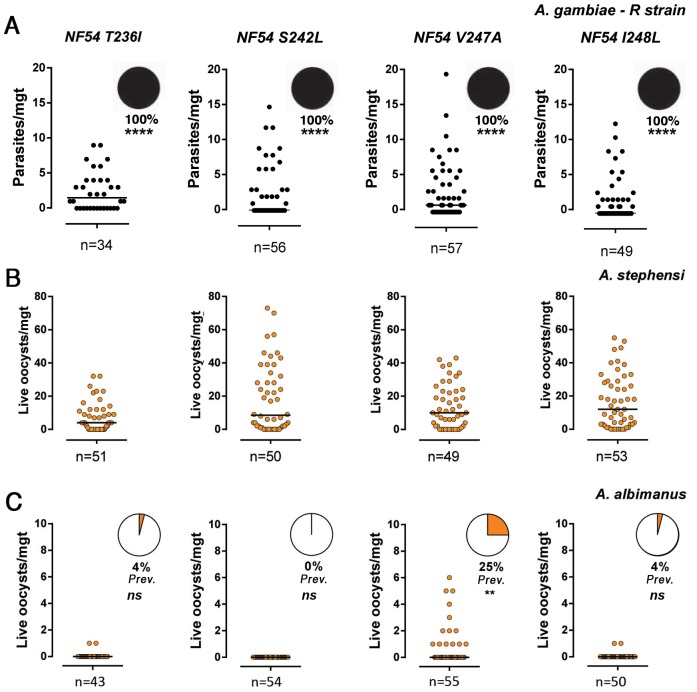
We confirmed that most (98%) WT NF54 parasites survive in A. gambiae R strain mosquitoes (Fig 1A, S1A Fig). In each experiment the viability of the gametocyte culture was confirmed by infecting A. stephensi (Nijmegen Sda500) mosquitoes, a strain that has been genetically selected in the laboratory to be highly susceptible to P. falciparum (Welch) (Fig 1A–1C, S1A–S1C Fig) [12]. In contrast to NF54 WT parasites, a P. falciparum Pfs47 KO NF54 line in which an attB/attP recombinase target site was included in the insertion that disrupts the gene [7] was completely (100%) melanized by A. gambiae R strain females (Fig 1B, S1B Fig). This is the same phenotype previously observed with an independent Pfs47 KO NF54 line [11]. The P. falciparum Pfs47 KO NF54 line was complemented with the NF54 Pfs47 haplotype (Pfs47 KO+NF54) using the attB/attP recombinase target site to generate a stable parasite line. Genetic complementation with the Pfs47 WT NF54 haplotype completely rescued parasite survival in the R strain (0% melanization) (Fig 1C, S1C Fig), confirming that the strategy was viable and that the stable transformants were functionally complemented.
Fig 1. Infection phenotype of different P. falciparum lines (NF54 WT, Pfs47 KO, Pfs47 KO + NF54) in the A. gambiae R strain midgut 7–9 d postfeeding.
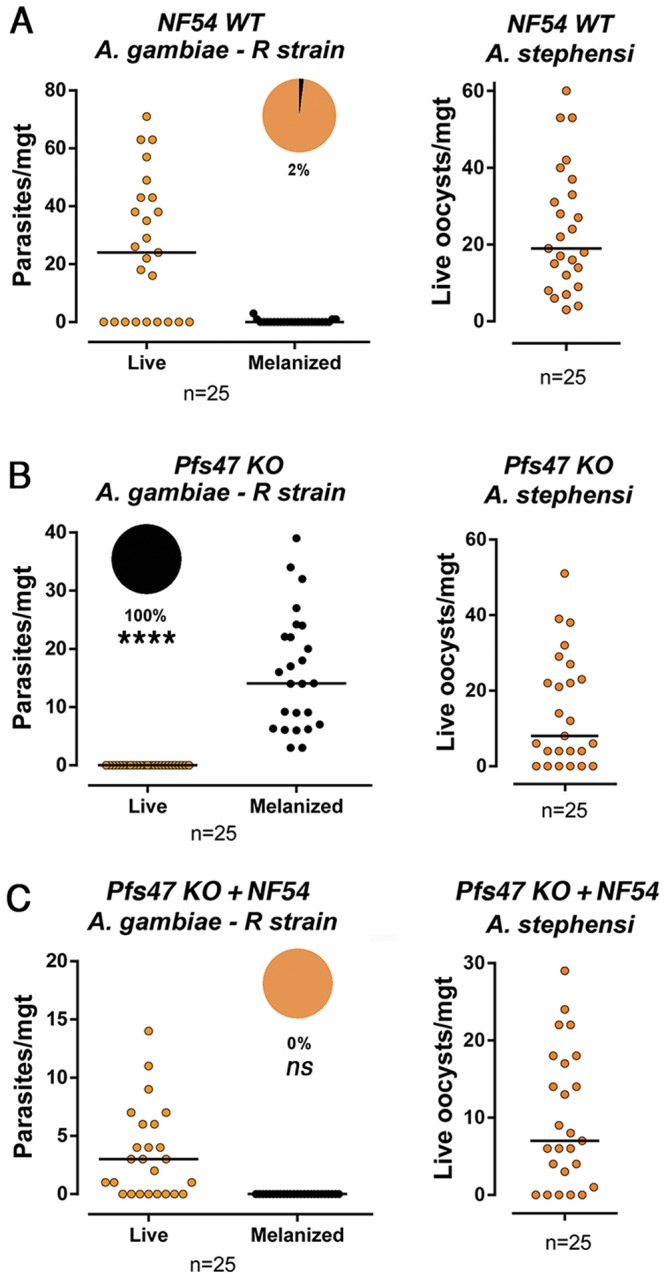
Live and melanized parasites on individual mosquito midguts in the A. gambiae R strain and confirmation of gametocyte viability in the susceptible A. stephensi Nijmegen using P. falciparum NF54 (A), Pfs47 KO (B) and Pfs47 KO + NF54 (C). The medians are indicated with black lines and proportion of live (orange) and melanized (black) parasites are indicated with pie charts. Each dot represents the number of parasite on an individual mosquito and the median is indicated with a black line (n = number of midguts examined). The differences in the proportion of melanized parasites relative to NF54 WT were analyzed using the X2 test, **** p<0.0001, ns = not significant. All parasite phenotypes were confirmed in two independent experiments (S1 Fig).
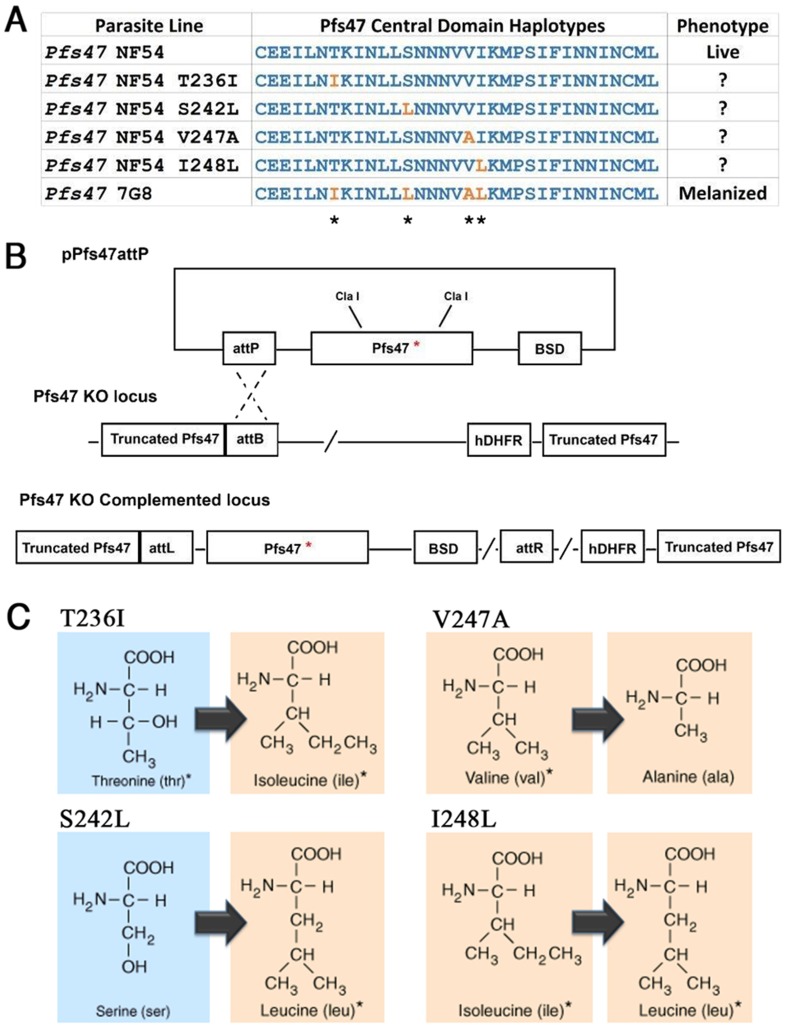
The molecular basis of Pfs47 phenotypic variation between the Africa and New World P. falciparum lines in the R strain was investigated by independently introducing each of the four single amino acid differences, one at a time, into the backbone of the previously reported pPsf47attP vector [7] using PCR-based mutagenesis (Fig 2A–2C). Following co-transfection of the pPfs47attP plasmids with an integrase-expressing plasmid and drug selection, the transgenic parasites complemented with the modified Pfs47 constructs T236I, S242L, V247A and I248L were recovered and cloned by minimal dilution. Gene complementation was confirmed by PCR-amplification and sequencing; and gene copy number, quantitative mRNA and protein expression were also verified for all the lines (S3 Fig). We expected that the first two mutations, T236I and S242L would have a strong phenotypic effect, as they replace polar amino acids with hydrophobic ones that also have bulkier side chains (Fig 2C). The other two mutations (V247A and I248L) are more conservative, as they do not alter the polarity of the side chains; and in the case of I248L the only difference is the position of a methyl group in the side chain (Fig 2C).
Fig 2.
(A) Schematic representation and alignment of Pfs47 Central Domain Haplotypes sequences used in this study. Variable positions between the NF54 haplotype and the 7G8 haplotype along with the point mutations constructs used in this study to complement the Pfs47KO line are marked as orange. The known infection phenotype of these strains in the A. gambiae R strain midgut is shown on the right. (B) Schematic representation of the integrase-mediated complementation of different Pfs47 haplotypes on the Pfs47KO locus by the plasmid pPfs47attP. The drug selection cassettes hDHFR and BSD the recombination adaptor sites attP/attB are showed. Representation is not in scale and is for illustration purposes only. (C) Schematic representation of the amino acids substituted on every single Pfs47 point mutation construct. Nucleophilic amino acids are shaded in cyan and hydrophobic amino acids are shaded in orange.
As expected, the two lines in which Pfs47 was mutated with a non-conservative single amino acid replacement (T236I and S242L) were no longer functionally complemented, and were unable to infect the A. gambiae R strain (100% melanized) (Fig 3A, S1 Table). We were surprised that the other two lines with more conservartive mutations (V247A and I248L) had a similar phenotype. The quality of the culture and the viability of the complemented lines was confirmed by infecting A. stephensi Sda500 mosquitoes side-by-side with the same cultures (Fig 3B, S3 Table). These results indicated that there is a strong specificity between the African Pfs47 protein of the parasite and the putative mosquito receptor, as any amino acid substitution, even a conservative one, disrupts the ability of WT NF54 Pfs47 to evade the immune system of the A. gambiae R strain.
Fig 3. Infection phenotype of different Pfs47 complement P. falciparum lines (T236I, S242L, V247A and I248L) in the A. gambiae R strain, A. albimanus and A. stephensi 7–9 d post-feeding representing two pooled independent experiments (Experiment 1 and Experiment 2, S1–S3 Tables).
(A) Infection of A. gambiae R strain. The pie charts indicate the proportion of live (orange) and melanized (black) parasites. (B) Infection of A. stephensi Nijmegen, and (C) A. albimanus mosquitoes with P. falciparum NF54 Pfs47KO complemented derivatives expressing Pfs47 haplotypes NF54 T236I, S242L, V247A and I248L. The orange area of the pie charts indicates the prevalence of infection in A. albimanus. Each dot represents the number of parasite on an individual mosquito and the median is indicated with a black line (n = number of midguts examined). Melanization prevalences in A. gambiae R strain were compared with the X2 test relative to Pfs47 KO + NF54, infection prevalences in A. albimanus were compared with the X2 test, ns = no significant difference,** p<0.01, **** p<0.0001. All parasite phenotypes were confirmed in two or three independent experiments.
We have previously shown that the Pfs47 KO line cannot infect A. albimanus mosquitoes (0% prevalence of infection), but introducing the American-7G8 Pfs47 haplotype (that differs from the African-GB4 line only in these four amino acids) greatly enhances infection reaching a prevalence of 63% in this New World vector. We tested whether introducing one amino acid change at a time could enhance infection in A. albimanus. In this mosquito the complement system lyses the parasites and no melanizations are observed. The S242L mutation had no effect (0% prevalence) (Fig 3C, S2 Table), the T236I and I248L substitutions had a very minor effect (2–4% prevalence, not significant, χ2 test), while the V247A mutation had a stronger effect enhancing infection and reaching a prevalence of infection of 25–30% (p<0.01, χ2 test, Figs 3C and 4, S2 Fig, S2 Table). We then tested whether the A. albimanus complement-like system was activated and limiting infection with two partially rescued lines. Disruption of the complement system by silencing LRIM1 greatly enhanced the intensity of infection with both lines (p< 0.0001, Fig 4, S2 Fig). The effect on the I248L lines was stronger, as the prevalence of infection increased from 5% in the LacZ control to 75% (37.5 fold) in the LRIM1-silenced group, while in the V247A line the prevalence increased from 30% to 80% (2.6 fold)(Fig 4, S2 Fig).
Fig 4. Effect of LRIM1 silencing in A. albimanus mosquitoes on its infection with P. falciparum Pfs47 NF54 V247A and I248L.
Effect of silencing the leucine-rich repeat immune protein 1 (LRIM1) in A. albimanus on infection with NF54 Pfs47 haplotypes V247A and I248L. Each dot represents the number of parasite on an individual mosquito and the median is indicated with a black line (n = number of midguts examined). The orange area of the pie charts indicates the prevalence of infection. All parasite phenotypes were confirmed in two independent experiments. Median number of oocysts were compared using the Mann–Whitney test. ****P < 0.0001.
Discussion
Global analysis of 364 P. falciparum isolates identified 42 different Pfs47 haplotypes with strong geographic structure and differences of compatibility between isolates from Africa, Asia, or the Americas and the mosquito vectors from these regions [7]. In general, parasites were more effective infecting mosquitoes from the same continent they originated from, suggesting that mosquito vectors select the parasites genotypes that circulate in a given geographic area. Furthermore, we demonstrated that incompatible parasites are eliminated by the mosquito complement-like system and that replacing the Pfs47 haplotype expressed in a parasite was sufficient to change the compatibility with a given vector. Based on these observations the “Lock and Key” model of Plasmodium mosquito immune evasion was proposed stating that: “We can think of Pfs47 as a “key” that allows the parasite to “turn off” the mosquito detection system by interacting with some mosquito receptor protein(s) (the lock). There are different haplotypes of this key, and the parasite needs to have the right key for the “lock” present in a given mosquito species to survive, continue to be transmitted, and become established in a new region” [7].
In this manuscript we explored how precise the fit of the Pfs47 “key” with the mosquito receptor has to be for the parasite to evade immunity. Our studies indicate that, in the A. gambiae R strain, only a perfect fit of four critical amino acids in the central domain would do, as any amino acid modification, even two different conservative single amino acid replacements completely disrupted the ability of the parasites to hide from the mosquito immune system and they were all destroyed. The V247A mutation is very interesting, because it greatly reduced parasite survival in the A. gambiae R strain, but had the opposite effect in A. albimanus and enhanced infection. This indicates that this is a critical amino acid and that the two additional methyl groups in the side chain of the valine residue were probably preventing an effective interaction with the putative Pfs47 receptor in A. albimanus. However, the effect of this modification on immune evasion was partial, as disrupting the mosquito complement-like system, by silencing LRIM1 in A. albimanus, greatly enhanced the level of infection, indicating that many parasites are still detected and destroyed by the mosquito immune system. It should be noted that when a NF54 KO line is complemented with the Pfs47 haplotype with the four amino acids present in the New World 7G8 line, the parasite becomes “invisible” to the mosquito and disrupting the complement system has no effect on the infection [7]. Furthermore, all the Pfs47 haplotypes identified so far in the New World share the four critical amino acids present in the 7G8 haplotype [7]. Taken together, our findings indicate that besides the V247A mutation, the other three amino acids are also required for complete immune evasion in A. albimanus.
We have previously shown that the JNK pathway is a major effector of mosquito antiplasmodial immunity, as it regulates the expression of the enzymes that mediate midgut epithelial nitration in response to Plasmodium invasion [13]; and epithelial nitration is necessary for effective activation of the mosquito complement-like system [14]. The R strain is in a chronic state of oxidative stress that is enhanced following a blood meal [15], and exhibits a constitutive level of systemic activation of JNK signaling with increased expression of the enzymes that mediate midgut nitration (NOX5 and HPX2) and of the antiplasmodial effectors TEP1 and FBN9 in hemocytes [13]. Furthermore, disruption of JNK signaling in the R strain greatly enhances infection with P. berghei, increasing the prevalence of infection from 0 to 68% [13].
A detailed functional analysis of the mosquito immune response to WT and Pfs47 KO P. falciparum (NF54 genetic background) revealed that Pfs47 allows the parasite to evade immunity by disrupting JNK signaling and preventing activation of epithelial nitration in response to ookinete midgut invasion [16]. The higher basal level of JNK activation in the R strain seems to pose a greater challenge to the parasite and may explain the observed results. A high affinity interaction with the mosquito Pfs47 receptor that triggers the disruption of JNK signaling appears to be required for P. falciparum to survive in mosquitoes with the R strain genetic background. Even minor modifications, such as the V247A or the I248L mutations, that could reduce the binding affinity, but are not predicted to have major effects on the overall molecular interaction, disrupt the parasite’s ability to block the mosquito antiplasmodial response. This level of specificity supports that idea that, in A. gambiae R strain, Pfs47 functions like a “key” that must fit the receptor and suggests that the region between the two cysteines in the D2 domain, where all the amino acid differences were present, is crucial for the interaction with the putative receptor in the mosquito midgut. A recent report identified an African P. falciparum line (NF165) in which the sequence of the D2 domain of Pfs47 is identical to that of NF54 and GB4 parasites, but most parasites (up to 66%) are melanized in the R strain [17]. Unfortunately, only 59% of the NF165 Pfs47 gene sequence was determined [17], and a recent global analysis of P. falciparum lines identified 11 non-synonymous polymorphisms in the region that was not sequenced [7]. It is thus possible that some polymorphism(s) are present in this region of the NF165 line (but were not sequenced) and this may be sufficient to disrupt the interaction with the receptor. Alternatively, a compatible Pfs47 haplotype may be necessary, but not sufficient for parasites to survive. Polymorphisms in other parasite genes present in the lines analyized by Eldering et al. [17] may also be critical for Plasmodium survival. The parasite may also require, for example, a higher capacity to detoxify reactive oxygen species (ROS), as the JNK-mediated induction of NOX5 expression generates high levels of intracellular ROS as the parasites traverse the mosquito midgut epithelial cell.
Some vectors seem to be more permissive, for example, the A. gambiae G3 strain can be infected with both NF54 and 7G8 parasites. However, it is more susceptible to NF54 infection than to 7G8 [7]. Conversely, A. albimanus is more susceptible to 7G8 than to NF54 [7]. The A. stephensi (Nijmegen Sda500) strain, that was genetically selected in the laboratory to be highly susceptible to P. falciparum (Welch), is highly susceptible to infection with all Pfs47 haplotypes tested so far, and is readily infected even with Pfs47 NF54 KO parasites (Fig 1B, S1B Fig). At the other end of the spectrum, in A. gambiae R strain, there is no room for variation in the four critical amino acids in the Pfs47 central domain and it may represent an extreme case in which only a perfect fit of Pfs47 with its receptor allows P. falciparum to survive. The goal of this study was to minimize as much as possible the genetic variation, both in the vector and parasite, that could influence the phenotypes, so that one could asses the effect of single amino acid differences on vector/parasite compatibility and transmission. The observed results with the anopheline laboratory lines used in this study cannot be generalized to all mosquitoes of a given species. However, the observation that the NF54 V247A mutation disrupts the ability of the parasite to evade immunity in the A. gambiae R strain and results in complete elimination, but has the opposite effect on A. albimanus, enhancing infection, supports the hypothesis that the “correct key” (Pfs47 haplotype) that the parasite needs is different between evolutionary distant anopheline vectors of malaria. Understanding these complex vector/parasite interactions, under natural field conditions, would require direct genetic analysis of the P. falciparum Pfs47 haplotypes present on individual naturally-infected anopheline mosquitoes and, ideally, simultaneous analysis of the mosquito Pfs47 receptor variant in the same sample. We conclude that malaria transmission involves a complex interplay between the genetic background of the parasite and the mosquito and that Pfs47 can be critical in this interaction, as it mediates Plasmodium immune evasion through molecular interactions that need to be very precise in some parasite/vector combinations.
Materials and Methods
Anopheles Mosquitoes and Plasmodium Parasites
Anopheles gambiae L35 (R strain), Anopheles albimanus and Anopheles stephensi Nijmegen Sda500 strains were used. Mosquitoes were reared at 27°C and 80% humidity on a 12-h light-dark cycle under standard laboratory conditions. The Plasmodium falciparum strains used—NF54, NF54-Pfs47KO and Pfs47 complemented lines (Africa-NF54, NF54 T236I, NF54 S242L, NF54 V247A and NF54 I248L)—were maintained in O+ human erythrocytes using RPMI 1640 medium supplemented with 25 mM HEPES, 50 mg/l hypoxanthine, 25 mM NaHCO3, and 10% (v/v) heat-inactivated type O+ human serum (Interstate Blood Bank, Inc., Memphis, TN) at 37°C and with a gas mixture of 5% O2, 5% CO2, and balance N2 [11].
Experimental Infection of Mosquitoes with P. falciparum
Mosquito females were infected artificially by membrane feeding with P. falciparum gametocyte cultures. Gametocytogenesis was induced as previously described [11]. Mature gametocyte cultures (stages IV and V) that were 14–16 d were used to feed mosquitoes using membrane feeders at 37°C for 30 min. Midguts were dissected 8 d after feeding, and oocysts were stained with 0.1% (wt/vol) mercurochrome in water and counted by light microscopy. Median numbers of oocysts were compared using the Mann-Whitney test; infection prevalences and the proportion of melanized parasties were compared with the X2 test. All parasite phenotypes were confirmed in two to three independent experiments.
dsRNA-Mediated Gene Knockdown
Individual female A. albimanus mosquitoes were injected 1–2 d postemergence as previously described [7]. Briefly, mosquitoes were injected with 69 nL 3 μg/μL dsRNA solution 3–4 d before receiving a Plasmodium-infected blood meal. dsRNA A. albimanus LRIM1 were produced using the MEGAscript RNAi Kit (Ambion, Austin, TX) using DNA templates obtained by PCR using A. albimanus cDNA and the primers previously described [7] with T7 polymerase promoter sites added in the 5ˊ-end. A. albimanus LRIM1 gene silencing was assessed in whole sugar-fed mosquitoes by quantitative real-time PCR using primers AaLRIM1-qF, AaLRIM1-q, respectively and were found to be 80% lower in dsRNA immune genes injected mosquitoes compared with a dsLacZ-injected control.
Stable Genetic Complementation of P. falciparum Pfs47KOattB
The generated Pfs47KOattB line was complemented with different point mutated alleles of NF54-Pfs47 with the plasmid pPfs47attP [7]. Due to the inability of generating point mutations in the 8.3 kb pPfs47attP, the domain two of the Pfs47 from P. falciparum NF54 was PCR amplified with the In-Fusion® designed oligonucleotides prClaF and prClaR and cloned into a pGemT-easy plasmid, generating pGemT-easy-Pfs47D2 plasmid. Point mutations were generated in the latter plasmid using the QuickChange Point Mutagenesis kit (Agilent) with the primers T236I_sense and T236I_antisense; S242L_sense and S242L_antisense, V247A_sense and V247A_antisense; I248L-sense and I248L_antisense, respectively. Then the point mutated fragment was PCR amplified with the primers prClaF and prClaR and introduced by infusion into the ClaI digested pPfs47attP plasmid to generating pPfs47attP NF54 T236I, NF54 S242L, NF54 V247A and NF54 I248L. Cloning procedures were carried out in TOP10F bacteria (Invitrogen) and every construct was fully sequenced (Operon).
P. falciparum Pfs47KO was cotransfected with a particular pPfs47attP (NF54 T236I, NF54 S242L, NF54 V247A or NF54 I248L) and the pINT—which encodes for the mycobacteriophage Bxb1 serine integrase in order to catalyze the integration of the plasmid pPfs47attP [18]—as previously describe with modifications [19]. Briefly, transfection mixture was prepared at room temperature and consisted of 20 μl packed RBCs, 5 μg of each plasmid DNA (1μg/ul) (Qiagen plasmid Maxiprep kit) and of 70 μl Amaxa SE solution resulting in a final volume of 100 μl. The transfection mixture was transferred to an Amaxa Nucleocuvette (Lonza) and transfection was performed at room temperature using an Amaxa 4D-Nucleofector. After applying the CM-162 pulse, electroporated RBC’s were processed as mentioned above. Culture media was changed daily and selection drugs (2.5 μg/ml Blasticidin HCl and 200μg/ml Geneticin) were added 24 hours after electroporation and maintained continuously in the asexual cultures unless stated otherwise. Once parasitemia was observed (day 21–28), parasites were cloned by minimal dilution in 96 wells plates (day 1). On day 14, 5ul aliquots of the 96 well plates were incubated with 2X SYBR-green-1 (Invitrogen) and 165 nM MitoTracker Deep Red (Invitrogen) in PBS and measure of live parasite were assessed using flow cytometry [7]. Positive wells were transferred to 24 well plates and later to 6 well plate.
Nucleic Acid Analysis
Primers pairs prIA1’F2/prIA1’R and prIA2’F/prIA2’R were used to detect both integration arms 1 and 2 respectively of the pPfs47attP’s plasmids into the Pfs47KO locus on chromosome 13. Pfs47 gene copy number was estimated by testing each DNA sample by qPCR with the primers PF13_0248F and PF13_0248R in parallel against the P. falciparum gene Pf10_0203 (ADP-ribosylation factor) gene using the primers 0203F and 0203R [7]. Pfs47 mRNA expression in the lines used in this work was confirmed by qPCR on cDNA from stage IV-V gametocyte cultures using the mentioned primers as previously described [7]. Estimation of gene dosage and mRNA expression was calculated according to the ΔΔCt method [20].
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank André Laughinghouse and Kevin Lee for insectary support.
Abbreviations
- APL1
Anopheles Plasmodium-responsive leucine-rich repeat 1 protein
- JNK
Janus N-terminal Kinase
- LRIM1
leucine-rich repeat immune protein 1
- TEP1
Thioester-containing Protein
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. World Malaria Report. Geneva: WHO, 2015. [Google Scholar]
- 2.Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–5. Epub 2010/09/25. 10.1038/nature09442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, et al. A global map of dominant malaria vectors. Parasit Vectors. 2012;5:69 Epub 2012/04/06. 10.1186/1756-3305-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molina-Cruz A, Barillas-Mury C. The remarkable journey of adaptation of the Plasmodium falciparum malaria parasite to New World anopheline mosquitoes. Memórias do Instituto Oswaldo Cruz. 2014;109(5):662–7. 10.1590/0074-0276130553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins FH, Sakai RK, Vernick KD, Paskewitz S, Seeley DC, Miller LH, et al. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science. 1986;234(4776):607–10. [DOI] [PubMed] [Google Scholar]
- 6.Molina-Cruz A, Dejong RJ, Ortega C, Haile A, Abban E, Rodrigues J, et al. Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of Anopheles gambiae mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 2012. Epub 2012/05/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina-Cruz A, Canepa GE, Kamath N, Pavlovic NV, Mu J, Ramphul UN, et al. Plasmodium evasion of mosquito immunity and global malaria transmission: The lock-and-key theory. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(49):15178–83. Epub 2015/11/26. 10.1073/pnas.1520426112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116(5):661–70. Epub 2004/03/10. [DOI] [PubMed] [Google Scholar]
- 9.Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, et al. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe. 2009;5(3):273–84. Epub 2009/03/17. 10.1016/j.chom.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 10.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;324(5924):258–61. Epub 2009/03/07. 10.1126/science.1171400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, et al. The Human Malaria Parasite Pfs47 Gene Mediates Evasion of the Mosquito Immune System. Science. 2013;340(6135):984–7. 10.1126/science.1235264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldmann AM, Ponnudurai T. Selection of Anopheles stephensi for refractoriness and susceptibility to Plasmodium falciparum. Medical and veterinary entomology. 1989;3(1):41–52. Epub 1989/01/01. [DOI] [PubMed] [Google Scholar]
- 13.Garver LS, de Almeida Oliveira G, Barillas-Mury C. The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLoS pathogens. 2013;9(9):e1003622 10.1371/journal.ppat.1003622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira GdA, Lieberman J, Barillas-Mury C. Epithelial Nitration by a Peroxidase/NOX5 System Mediates Mosquito Antiplasmodial Immunity. Science. 2012;335(6070):856–9. 10.1126/science.1209678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Christophides GK, Cantera R, Charles B, Han YS, Meister S, et al. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):14139–44. 10.1073/pnas.2036262100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramphul UN, Garver LS, Molina-Cruz A, Canepa GE, Barillas-Mury C. Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proceedings of the National Academy of Sciences. 2015;112(5):1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eldering M, Morlais I, van Gemert G-J, van de Vegte-Bolmer M, Graumans W, Siebelink-Stoter R, et al. Variation in susceptibility of African Plasmodium falciparum malaria parasites to TEP1 mediated killing in Anopheles gambiae mosquitoes. Scientific reports. 2016;6:20440 10.1038/srep20440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adjalley SH, Lee MC, Fidock DA. A method for rapid genetic integration into Plasmodium falciparum utilizing mycobacteriophage Bxb1 integrase. Methods Mol Biol. 2010;634:87–100. Epub 2010/08/03. 10.1007/978-1-60761-652-8_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caro F, Miller MG, DeRisi JL. Plate-based transfection and culturing technique for genetic manipulation of Plasmodium falciparum. Malar J. 2012;11:22 10.1186/1475-2875-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001. December;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.