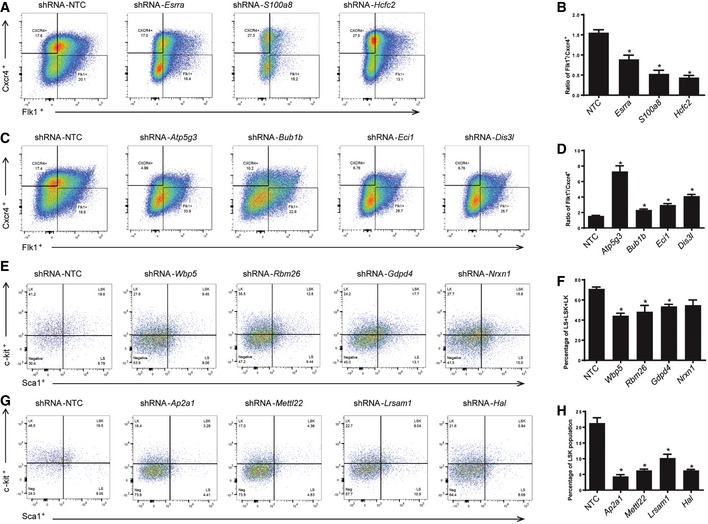
The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Esrra, S100a8, or Hcfc2 and induced to form EBs. The day 6 EB cells were analyzed by flow cytometry to measure the mesoderm/endothelium cells (Flk1+/Cxcr4−) and endoderm cells (Cxcr4+/Flk1−). The percentages of mesoderm/endothelium cells (Flk1+/Cxcr4−) were decreased on day 6 with shRNA knockdown of Group XII genes Esrra, S100a8, and Hcfc2 compared to control as quantified by flow cytometric analysis.
Knockdown of Esrra, S100a8, and Hcfc2 showed that the ratios of percentage of mesoderm/endothelium population vs. endoderm population were decreased as measured by Flk1+/Cxcr4− vs. Flk1−/Cxcr4+ on day 6. The data were generated based on the quantification of flow cytometric analyses. Error bars represent standard deviation (SD), n = 2. Student's t‐test, *P < 0.05.
The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Atp5g3, Bub1b, Eci1, or Dis3l and induced to form EBs. The day 6 EB cells were analyzed by flow cytometry to measure the mesoderm/endothelium cells (Flk1+/Cxcr4−) and endoderm cells (Cxcr4+/Flk1−). Flow cytometric analysis showed that endoderm cells (Cxcr4+/Flk1−) were deceased while mesoderm/endothelium cells (Flk1+/Cxcr4−) were slightly increased in the cells with shRNA knockdown of Group XI genes Atp5g3, Bub1b, Eci1, and Dis3l compared to control.
Knockdown of Atp5g3, Bub1b, Eci1, or Dis3l led to increase of mesoderm/endothelium/endoderm population ratio as quantified based on the flow cytometry data. Error bars represent standard deviation (SD), n = 3. Student's t‐test, *P < 0.05.
The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Wbp5, Rbm26, Gdpd4, and Nrxn1 and differentiated toward HSPCs. The differentiated cells on day 20 were analyzed by flow cytometry. Silencing of Group IV genes including Wbp5, Rbm26, Gdpd4, and Nrxn1 decreased overall LK, LS, and LSK population on day 20.
Quantification of LK + LSK + LS cell fractions with Wbp5, Rbm26, Gdpd4, and Nrxn1 silencing based on flow cytometric analyses. Error bars represent standard deviation (SD), n = 2. Student's t‐test, *P < 0.05.
The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Ap2a1, Mettl22, Lrsam1, or Hal and differentiated toward HSPCs. The differentiated cells on day 20 were analyzed by flow cytometry. Flow cytometric analysis showed notable reduction of LSK population on day 20 with Group X genes Ap2a1, Mettl22, Lrsam1, or Hal knockdown.
Quantification of flow cytometric analyses showed knockdown of Ap2a1, Mettl22, Lrsam1, and Hal significantly impaired generation of LSK population on day 20 in vitro compared to control. Error bars represent standard deviation (SD), n = 3. Student's t‐test, *P < 0.05.