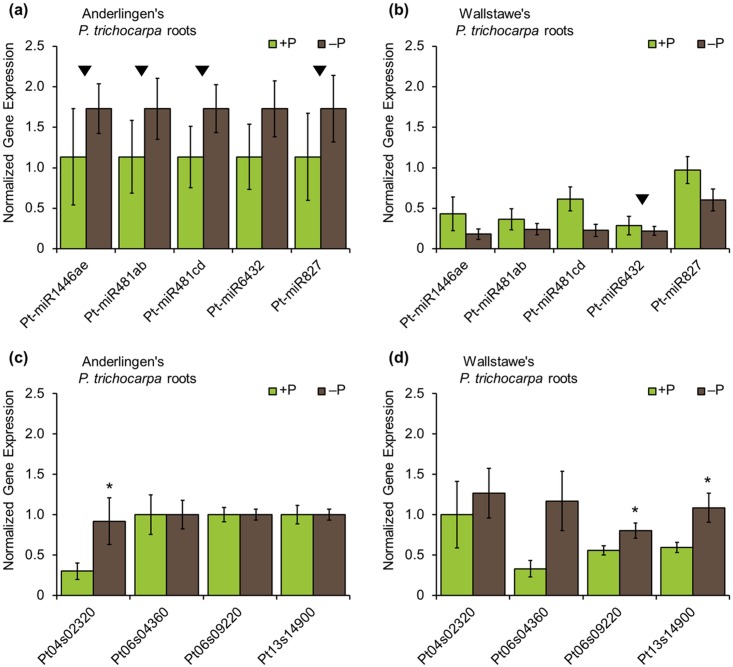
Fig 7. Target gene expression differences of clonal Populus trichocarpa material.
qPCR results from Populus trichocarpa (cv. Muhle Larson) root material derived from two different short rotation forestry sites (Anderlingen (a,c) and Wallstawe (b,d)), grown under controlled adequate (+P) vs. deficient (−P) phosphorus nutrition using 3 reference genes (POPTR_EF103B0031, POPTR_RP and POPTR_18s) for normalization. Normalized gene expression (y-axis) is shown for differentially methylated miRNAs (a,b) and their possible target genes (c,d): Ptc-miR1446ae, Ptc-miR481ab, Ptc-miR4b1cd, Ptc-miR6432, Ptc-miR827, POPTR_0004s02320 as Pt04s02320, POPTR_0006s04360 as Pt04s04360, POPTR_0006s09220 as Pt06s09220 and POPTR_0013s14900 as Pt13s14900 (x-axis). Black triangles indicate which plant material had a higher methylation level. Data are presented as the mean ± SEM, p* ≤ 0.05 and 95% confidence intervals and were obtained from 3 independent experiments.