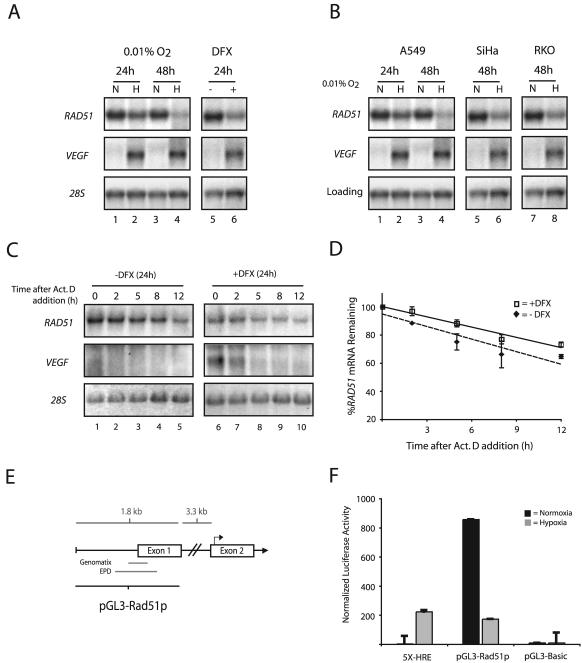
FIG. 3.
Transcriptional repression of the RAD51 gene by hypoxia. (A) Northern blot analyses were performed on total RNA extracted from MCF-7 cells after exposure to normoxia (lanes N), hypoxia (0.01% O2) (lanes H), or DFX (250 μM). The time for which cells were maintained under each condition (24 or 48 h) is given. VEGF expression is shown for comparison, to verify that physiologically relevant levels of hypoxia were present in the treated cells, and expression of 28S rRNA is presented to confirm equal sample loading. (B) Northern blot analysis of RAD51 mRNA expression in A549, SiHa, and RKO cells after a 24- or 48-h exposure to hypoxia (0.01% O2). VEGF expression is shown for comparison, to verify that physiologically relevant levels of hypoxia were present in the treated cells. Expression of 28S rRNA (MCF-7, SiHa, and RKO cells) and β-actin mRNA (A549 cells) is presented to confirm equal sample loading. (C) To assess the stability of RAD51 mRNA, MCF-7 cells were either left untreated or exposed to DFX (250 μM) for 24 h, followed by coincubation with ActD (5 μg/ml) to block transcription. Cells were harvested at the indicated times after the addition of ActD, and RAD51 mRNA expression was determined by Northern blotting. Expression of VEGF is shown to confirm both the induction of chemical hypoxia and successful abolition of transcription. In addition, 28S rRNA levels were unchanged and served as standards to confirm equal sample loading. (D) Analysis of RAD51 mRNA expression at each time point after ActD addition in cells exposed to DFX or left untreated, as determined by phosphorimager analysis of Northern blots. Values are the percentage of RAD51 mRNA remaining in either DFX-treated or untreated cells at each time point, and error bars are based on standard errors calculated from duplicate experiments. (E) Schematic of the 5′-flanking region of the RAD51 gene, with delineation of the promoter fragment used for luciferase reporter gene assays (pGL3-Rad51p). Approximate locations of the core promoter regions, as described in the Eukaryotic Promoter Database (EPD) and as identified by in silico analysis using the Genomatix promoter identification algorithm PromoterInspector, are shown for reference. Bent arrow above exon 2 indicates the ATG translation start codon. (F) To determine the effect of hypoxia on RAD51 gene promoter activity, the pGL3-Rad51p luciferase (firefly) reporter plasmid was transiently transfected into RKO cells 4 h prior to normoxic or hypoxic exposure (for 48 h), immediately followed by measurement of luciferase activity. Firefly luciferase values were normalized to Renilla luciferase activity from a cotransfected pRL-SV40 control vector, and error bars are based on standard errors calculated from duplicate experiments. The activity of the luciferase reporter plasmid 5X-HRE, which contains five HREs tandemly ligated to a human cytomegalovirus minimal promoter, is shown as a control to confirm physiologically relevant levels of hypoxia. The activity of the promoterless luciferase reporter gene construct pGL3-Basic is also shown as a control.