Abstract
Tumor necrosis factor alpha (TNF-α)-related apoptosis-inducing ligand (TRAIL) is a member of the TNF-α family of death receptor ligands and holds great therapeutic potential as a tumor cell-specific cytotoxic agent. Using a panel of established tumor cell lines and normal cells, we found a significant difference between the number of TRAIL-sensitive cells expressing high levels of c-myc and TRAIL-resistant cells expressing low levels of c-myc (P < 0.05, n = 19). We also found a direct linear correlation between c-myc levels and TRAIL sensitivity in TRAIL-sensitive cell lines (r = 0.94, n = 6). Overexpression of c-myc or activation of a myc-estrogen receptor (ER) fusion sensitized TRAIL-resistant cells to TRAIL. Conversely, small interfering RNA (siRNA)-mediated knockdown of c-myc significantly reduced both c-myc expression and TRAIL-induced apoptosis. The gene encoding the inhibitor of caspase activation, FLICE inhibitory protein (FLIP), appears to be a direct target of c-myc-mediated transcriptional repression. Overexpression of c-myc or activation of myc-estrogen receptor (ER) decreased FLIP levels both in cell culture and in mouse models of c-myc-induced tumorigenesis, while knocking down c-myc using siRNA increased FLIP expression. Chromatin immunoprecipitation and luciferase reporter analyses showed that c-myc binds and represses the human FLIP promoter. c-myc expression enhanced TRAIL-induced caspase 8 cleavage and FLIP cleavage at the death-inducing signaling complex. Combined siRNA-mediated knockdown of FLIP and c-myc resensitized cells to TRAIL. Therefore, c-myc down-regulation of FLIP expression provides a universal mechanism to explain the ability of c-myc to sensitize cells to death receptor stimuli. In addition, identification of c-myc as a major determinant of TRAIL sensitivity provides a potentially important screening tool for identification of TRAIL-sensitive tumors.
The cell surface death receptor ligand tumor necrosis factor alpha (TNF-α)-related apoptosis inducing ligand (TRAIL) can selectively kill cancer cells while leaving most normal cells intact (3, 20). While other members of the TNF family of death receptor ligands considered for cancer therapy (e.g., TNF-α and CD95L/FasL) exert significant toxicity, preclinical studies using TRAIL appear encouraging (4, 56, 64). Recombinant TRAIL elicits little to no overt toxicity when administered systemically (4, 56, 64). However, human hepatocytes appear susceptible to TRAIL-induced apoptosis in vitro (29, 52). To circumvent this problem, other ways of activating TRAIL signaling have been suggested (26), as well as using TRAIL in combination with other drugs that may prevent hepatotoxicity (52). Variability in the preparations of recombinant TRAIL may also contribute to selective hepatotoxicity (37, 64).
As with most promising cancer therapies, TRAIL is not universally lethal toward all cancer cells. Questions about what makes a cancer cell susceptible to TRAIL has led to discoveries about what controls TRAIL action (2). TRAIL kills by binding one of two cell surface receptors, death receptor 4 (DR4, also known as APO2 and TRAILR1) or death receptor 5 (DR5, also known as TRAILR2, KILLER/DR5, and TRICK2). After binding TRAIL, these transmembrane receptors each assemble a death-inducing signaling complex (DISC): the DRs form homotrimers that signal through an adaptor protein, FADD, which recruits the apoptosis-initiating proteases caspase 8, which then self-activates and initiates a signaling cascade leading to apoptosis.
We have previously investigated which components of the TRAIL pathway are the key regulators of TRAIL resistance and sensitivity (11, 33). During the course of our studies, we have identified several cell lines that are either highly resistant or sensitive to killing by TRAIL. Because c-myc is known to sensitize cells to many death stimuli, including TNF-α and CD95/Fas, we examined whether cells that were sensitive to TRAIL also had high c-myc expression (25, 34). Here we present data showing a direct correlation between TRAIL sensitivity and constitutive levels of c-myc expression. We tested this relationship and found that knocking down c-myc expression in sensitive cells diminished TRAIL action and expressing c-myc in resistant cells sensitized them to TRAIL. We also identified FLICE inhibitory protein (FLIP), also known as Casper, I-FLICE, CASH, FLAME-1, MRIT, CLARP, and usurpin, as a major regulator of c-myc sensitization to TRAIL. FLIP is structurally related to caspase 8—it contains tandem death effector domains that bind to caspase 8 at the DISC and can block its activation (35). Here we show that c-myc represses FLIP transcription by binding to the FLIP gene promoter. These findings suggest that elevated c-myc expression is important in mediating TRAIL action by repressing FLIP transcription and that c-myc may be a potentially useful tumor-specific marker for identifying TRAIL-sensitive tumors.
MATERIALS AND METHODS
Cell lines and culture conditions.
WI38 normal human embryonic lung fibroblasts and the human cancer cell lines SkBr3, MCF-7, HCC1937, and BT549 (breast), U2OS and SAOS2 (osteosarcoma), SW480, DLD1, and HT29 (colon), CaLU6 (lung), DU145 and PC3 (prostate), OVCAR3, SKOV3, and HeLa (ovarian), HEPG2 (liver), ACHN (renal), and FADU (human nasopharyngeal cancer) were obtained from the American Type Culture Collection (Manassas, Va.) and cultured under the recommended conditions. H460 human lung cancer cells and HCT116 human colon cancer cells were cultured as previously described (67). Normal human foreskin fibroblasts (HFF) were the kind gift of Meenhard Herlyn (Wistar Institute). myc-estrogen receptor (ER)-expressing cells were generated by retroviral infection using pBabepuro3:MycER of early-passage WI-38 and HFF cells and U2OS and SkBr3 cancer cells and selected with 1 μg of puromycin per ml for 2 weeks. Pooled stable transfectants were used for analysis. Cells were treated with 500 nM 4-hydroxytamoxifen (Sigma) to activate the myc-ER fusion protein. For serum deprivation experiments, cells were incubated in 0.1% fetal calf serum-containing medium for 48 h before being subjected to adenovirus infection and harvested for analysis at the indicated times postinfection. SkBr3 cells were treated with 50 ng of phorbol 12- myristate (Sigma) per ml for 20 h prior to the indicated treatment to arrest cells in the G1 phase of the cell cycle and rapidly reduce endogenous levels of c-myc (7, 46, 47). For TRAIL treatments, unless stated otherwise, cells were exposed to 50 ng of His-tagged recombinant human soluble TRAIL (Alexis, San Diego, Calif.) per ml plus 1 μg of anti-6x-histidine antibody (R&D Systems, Minneapolis, Minn.) per ml for 6 h prior to collection. Stable U2OS cells expressing tetracycline-inducible FLIP were generated by first introducing a regulatory element tetracycline-controlled transactivator by retroviral infection (pRevTet- On; Clontech) and selecting with G418. Pooled stable clones were then transduced with either an empty vector or a FLIP expression vector activated via tetracycline (pRevTRE; Clontech) by retroviral infection and selected with hygromycin.
Plasmid constructs.
The pCDNA3-cmyc expression vector for human c-myc was constructed by removing the previously cloned c-myc cDNA from pCRII-c-myc (46) using HindIII and XbaI and inserting it into pCDNA3 (Invitrogen). The myc-ER- expressing retroviral vector was generated by inserting human c-myc with a destroyed stop codon into the pBabepuro3:hbERTAM vector to translate in frame with the tamoxifen- sensitive hormone binding domain of the estrogen receptor (the kind gift of M. McMahon [14, 66]), as done previously by others (38). A 1,460-bp sequence containing the promoter region of the FLIP gene was amplified using the BAC clone RP11-536118 (Children’s Hospital Oakland Research Institute, Oakland, Calif.) and inserted into the luciferase-expressing reporter plasmid pGL3-basic (Promega). The sequence corresponds to positions −1179 to +281 relative to the proposed transcriptional start site of CFLAR (FLIP) exon 1 (accession no. AB038965 [23]). Two additional reporter plasmids were made by PCR amplification of the sequences corresponding to positions −503 to +281 and −503 to +102. Miz-1 cDNA was fused with the Flag epitope by PCR amplification of Miz-1 from plasmid pUHD-Miz-1 (the generous gift of M. Eilers), inserted into pCMV2-Flag (Sigma). YY1 cDNA was obtained from the American Type Culture Collection fused with the Flag epitope by PCR amplification, and inserted into pCMV2-Flag. Two DNA sequences that generate short hairpin small interfering RNA (siRNA) against FLIP were inserted into pSUPER.retro.gfp-neo plasmid (OligoEngine, Seattle, Wash.). They begin at positions 139 and 548 of the FLIP cDNA sequence (accession no. NM_003879).
Western blotting and antibodies.
Immunoblotting was carried out using antibodies against the following: c-myc (N-262; Santa Cruz Biotechnology, Santa Cruz, Calif.), human FLIP (NF-6; Axxora LLC, San Diego, Calif.), mouse FLIP (Dave-2; Axxora LLC), poly(ADP-ribose) polymerase (PARP) (Roche Molecular Diagnostics), Bcl-XL (556361; Pharmingen/BD Biosciences); Bcl-2 (Dako), caspase 8 (3-1-9; Pharmingen), caspase 10 (4C1; MBL, Woburn, Mass.), FADD (A66-2; Pharmingen), and caspase 3 (E-8; Santa Cruz Biotechnology). For loading control, a Ran antibody (Pharmingen/BD Biosciences) was used. Unless otherwise noted, immune complexes were detected by chemiluminescence and visualized using film. Proteins detected by chemiluminescence were quantified using a GelDoc 2000 imaging system with Quantity One software (Bio-Rad, Hercules, Calif.). To quantify c-myc protein, the immunoblots were incubated with fluorescently labeled secondary antibodies (Alexa-Fluor-680 anti-rabbit to detect c-myc [Molecular Probes], and IRDye800-labeled anti-mouse to detect Ran [Rockland]). An Odyssey infrared imaging system (LiCor, Lincoln, Neb.) was used to scan the immunoblot and quantify proteins. For immunohistochemistry of mouse tumor sections, rabbit anti-FLIP (LabVision, Fremont, Calif.) was used.
Adenovirus infections.
A human c-myc-expressing adenovirus that coexpresses green fluorescent protein (Ad-cMyc-GFP) and an adenovirus expressing green fluorescent protein alone (Ad-GFP) were generated using the Ad-Easy System (24). Adenovirus titers and infections were carried out as described previously (55).
Real-time quantitative RT-PCR assay.
Total RNA was prepared using the RNeasy minikit (Qiagen, Valencia, Calif.). TaqMan real-time quantitative reverse transcription-PCR (RT-PCR) was conducted as previously described using an Applied Biosystems 7700 sequence detector (10). FLIP and GAPDH PCR amplifications were performed concurrently in the same well. The following primer and probe sequences were used: FLIP primers, 5′-CACCGAGACTACGACAGCTTTGT-3′ and 5′-GCCCTGAGTGAGTCTGATCCA-3′; FLIP probe, 5′-FAM-CATACACACTCTGGGAGCCTCCTCGG-TAMRA-3′; GAPDH primers, 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′; GAPDH probe, 5′-VIC-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′.
Apoptosis assays. (i) Activated caspase 3 analysis.
The percentage of cells undergoing apoptosis was determined by measuring the active form of caspase 3 using flow cytometry. Cells were collected and fixed using the Cytofix/Cytoperm kit from Pharmingen. The fixed cells were suspended in 50 μl of Perm/Wash buffer. An antibody that specifically recognizes the active form of caspase 3 and not the proenzyme (C92-605; BD Biosciences) was added to a final concentration of 0.25 μg/sample. After a 20-min incubation and washing in Perm/Wash buffer, anti-rabbit Alexa-Fluor-647 R-phycoerythrin (Molecular Probes) was added to the cells (0.25 mg/50 ml). The cells were washed, and cells staining for GFP and Alexa-Fluor were counted. Experiments were performed in triplicate and repeated at least twice.
(ii) Propidium iodide staining.
In separate experiments, cells stained with prodium iodide were analyzed by flow cytometry for sub-G1 DNA content as described previously (51).
(iii) Annexin V staining.
Cells were collected and incubated with red-shifted phycoerythrin-conjugated recombinant human annexin V (CalTag, Burlingame, Calif.), and fluorescence was analyzed by flow cytometry using a Coulter-Beckman Elite Epics sorter.
Luciferase assays.
U2OS cells were plated in six-well plates and transfected using 4 μl of Lipofectamine2000 reagent (Invitrogen) plus a total of 4 μg of DNA. In all experiments, 0.1 μg of pCMV-β-galactosidase plasmid was added to measure transfection efficicency. In all experiments, 0.1 μg of pGL3-FLIP promoter-luciferase reporter plasmid or pGL3-basic was added. In experiments investigating c-myc, 3.2 μg of pCDNA3-c-myc or pCDNA3 was added. In other experiments, 1.6 μg of pFlag-Miz-1, pFlag-YY1, or pFlag, with 1.6 μg of pCDNA3-c-myc or pCDNA3, was used. At 24 hr after transfection, the cells were collected and luciferase activity was measured using the Luciferase assay system (Promega). Light units were normalized to β-galactosidase activity to control for transfection efficiency. There were no unexpected variations in β-galactosidase activity under the various transfection conditions. Results shown are from at least three separate experiments.
Chromatin immunoprecipitation.
A total of 3 × 106 U2OS cells were plated in a T75 flask, serum starved for 2 days, and infected with control or c-myc-expressing adenovirus for 24 h. The chromatin immunoprecipitation assay was performed as previously described (40) with a second immunoprecipitation step as described previously (68) and the following modifications. Chromatin was incubated with 10 μg of anti-c-myc antibody (N-262; Santa Cruz) or rabbit immunoglobulin (IgG) overnight, and immunocomplexes were collected by incubation for 4 h with protein-A/G agarose beads (Invitrogen). After being washed, the protein-DNA complexes were eluted with two successive incubations in 100 μl of 10 mM Tris-HCl (pH 8.0)-1 mM dithiothreitol-0.5% sodium dodecyl sulfate for 10 min. Iodoacetamide was added to a final concentration of 5 mM for 15 min to neutralize the dithiothreitol. Samples were diluted with 700 μl of dilution buffer (40), and the immunoprecipitation was repeated. After the DNA-protein complexes were eluted, the following were added: iodoacetamide (5 mM for 15 min), RNase A (50 μg/ml for 15 min at 37°C), and proteinase K (200 μg/ml for 30 min at 45°C). Cross-links were reversed by heating at 65°C for 6 h. DNA was phenol-chloroform extracted twice, chloroform extracted once, and ethanol precipitated. PCR amplification was performed using FastStart Taq polymerase (Roche). The primer pair used for amplification of the FLIP promoter corresponds to positions +159 to +300 relative to the proposed transcriptional start site (23). The primer sequences were as follows: forward primer, 5′- GTGTAGGAGAGAAGCGCCGCGAAC-3′; reverse primer, 5′-GGACTCTCCTGCCGCTGCCACCTC-3′. The primers used to amplify c-myc binding sites of the CAD gene were as follows: forward primer, 5′-TCTCTGCTGCTGCCGCCAAG-3′; reverse primer, 5′-ACCGACCCGTCCTCCAACAC-3′.
Transfection of siRNA oligonucleotides and pSUPER constructs.
The c-myc double- stranded SMARTPOOL siRNA oligonucleotides and double-stranded siRNA oligonucleotides corresponding to firefly luciferase (19) were purchased from Dharmacon Research (Lafayette, Colo.). Oligofectamine (Invitrogen) was used to transfect H460 and SW480 cells and Lipofectamine2000 was used to transfect HCT116 cells with siRNA oligonucleotides and plasmid DNA as specified by the manufacturers.
Mouse models of tumorigenesis. (i) Constitutive c-myc expression model.
Ki-ras-transformed p53−/− murine colonocytes were cultured as described previously (58). myc-overexpressing cells were derived by infection of Ki-ras colonocytes with a pMIGR1 retrovirus (53) encoding human c-myc together with enhanced GFP (EGFP) and purified by cell sorting. For tumor studies, 2 × 106 Ki-ras- or Ki-ras/c-myc-expressing colonocytes were injected subcutaneously into syngeneic C57BL6/J mice (The Jackson Laboratory, Bar Harbor, Maine). Animals were sacrificed after 15 days, and tumors were either fixed in buffered formalin or rapidly frozen for protein extraction. Serial 5-μm paraffin sections were prepared. Deparaffinized sections were heated for 15 min in 0.01 M citrate buffer (pH 6.2) in a microwave oven for antigen retrieval. The sections were then immunostained by the avidin-biotinylated enzyme complex peroxidase method (Vector Labs, Burlingame, Calif.) and subjected to a weak hematoxylin counterstain. For Western blotting, frozen tumor tissues were ground to powder by using a mortar and pestle. Proteins were extracted in RIPA buffer on ice for 30 min. After the protein content was determined using the DC protein assay kit (Bio-Rad), lysates were mixed with 2× Laemmli buffer and boiled for 5 min.
(ii) Conditional c-myc expression model.
myc-ER-induced hematopoietic neoplasms were generated as described previously for constitutive myc-induced tumors (69, 70). myc-ER was activated by daily intraperitoneal injections of 1 mg of 4-hydroxytamoxifen (Sigma) resuspended in pharmacy- grade olive oil as described previously (50). Total tumor RNAs were isolated, and reverse transcription of mRNA was performed using standard procedures. Real-time PCR analysis was performed by measuring SYBR Green incorporation using a LightCycler (Roche Diagnostics GmbH). The primers used to detect the mouse FLIP cDNA were 5′-CCACATCCGTGAAGAGACTTAC-3′ and 5′-TCCAAGGAGAACCCTGAGTGAAC-3′ and actin 5′-TTCGTTGCCGGTCCACA-3′ and 5′-ACCAGCGCAGCGATATCG-3′. Results are presented as relative mRNA levels (Ct) expressed as 2[30−Ct], where 2 is the assumed efficiency of PCR amplifications and 30 is the total number of cycles performed.
TRAIL DISC immunoprecipitation.
WI38-Myc-ER cells were suspended by treatment with trypsin and then treated with 1 μg of anti-6x-histidine antibody per ml with or without 100 ng of His-tagged recombinant human soluble TRAIL per ml for 30 min. Ice-cold phosphate-buffered saline (4 ml) was added to stop the reaction, and cells were collected by centrifugation (270 × g) at 4°C. The cells were washed in phosphate-buffered saline again and collected. They were lysed in 1 ml of lysis buffer (30 mM Tris-Cl [pH 7.5], 150 mM NaCl, 10% glycerol, 1 % Triton X-100, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Complete Mini; Roche]) for 30 min on ice, and debris was pelleted by microcentrifugation for 15 min at maximum speed. The supernatant was collected, and 30 μl was removed for input analysis. Protein G-conjugated agarose beads (30 μl of a 50% slurry) were added to immunoprecipitate DISC overnight by using end-over-end rotation. The beads were washed five times with lysis buffer, and protein complexes were eluted by addition of Laemmli sample buffer. Samples were boiled for 5 min, and proteins were separated by sodium dodacyl sulfate-polyacrylamide gel electrophoresis. Isotype-specific horseradish peroxidase secondary antibody (mouse IgG1; Southern Biotechnology Associates) was used in Western blottings to avoid cross-reacting bands.
RESULTS
TRAIL sensitivity correlates with high c-myc levels.
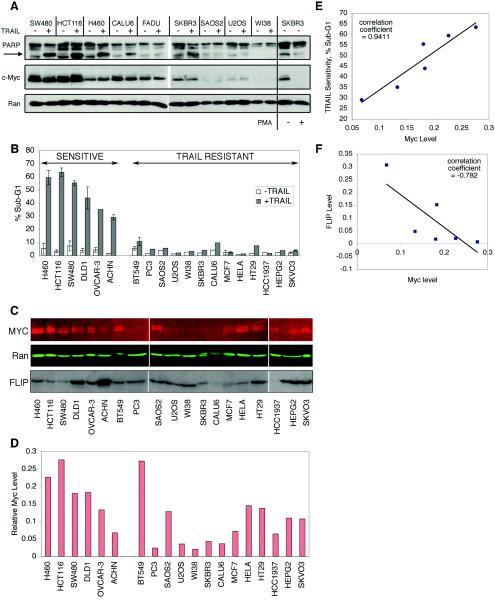
Because c-myc is known to sensitize cells to signaling through the TNF-α (34) and CD95/Fas (25) receptors, we hypothesized that endogenous c-myc levels may contribute to TRAIL sensitivity. We have shown previously that some cancer cell lines are sensitive to TRAIL while others are resistant (33, 51). We screened a panel of nine human cell lines, including cancer as well as normal diploid human fibroblasts (WI-38), for TRAIL-induced cell death (Fig. 1A). Exposing these cells to TRAIL for 6 h resulted in rapid death of the SW480, HCT116, and H460 cells, as shown by PARP cleavage. FADU cells are resistant due to a homozygous deletion in the DR4 gene and were excluded from further analysis (51). We noticed a potential correlation between the levels of c-myc and sensitivity to TRAIL. To further examine and quantify whether a statistically significant correlation existed between c-myc and TRAIL sensitivity, we increased our panel to include 19 cell lines, attempting to add more TRAIL-sensitive cells and cells expressing high c-myc levels (Fig. 1B to D). Of the 19 cell lines, 6 showed significant sensitivity to TRAIL (at least 20% cells undergo apoptosis after a 6-h exposure to 50 ng of TRAIL per ml [Fig. 1B]). To accurately determine c-myc protein levels, we used an infrared imaging system to quantify proteins detected by fluorescently labeled secondary antibodies. Figure 1C shows the c-myc immunoblot, and Fig. 1D shows the quantification of c-myc protein normalized to the Ran loading control for the 19 cell lines shown in Fig. 1B. We performed two statistical analyses to test whether c-myc levels correlate with TRAIL sensitivity. First, we applied a chi-square analysis, positing that c-myc expression and TRAIL sensitivity are independent. We observed a significant difference between the number of TRAIL-sensitive cells expressing high c-myc levels and TRAIL-resistant cells expressing low c-myc levels (χ2 = 4.59, P = 0.032, if high c-myc is defined as the level above average, and χ2 = 9.97, P = 0.0016, if high c-myc is defined as the level above the average plus 1 standard deviation). Because TRAIL-resistant cells may have defects in the death receptor signaling pathway (e.g., FADU cells [51]) and because it is unclear if some of the TRAIL-resistant cells in our cell panel possess such defects, we used a second test to determine if there is a significant relationship between c-myc and TRAIL sensitivity. We examined the TRAIL-sensitive cell lines for a correlation with the c-myc level and observed a direct linear relationship between the two variables, with a strong correlation coefficient (r = 0.941 [Fig. 1E]). We conclude from these data that (i) there is a strong probability that cells with above average c-myc levels (as defined using the cell panel shown in Fig. 1 as >0.118% Ran expression) will be sensitive to TRAIL and (ii) a direct linear relationship exists between c-myc levels and the amount of TRAIL-induced death in TRAIL-sensitive cells.
FIG. 1.
TRAIL sensitivity correlates with endogenous levels of c-myc protein. (A) The indicated cell lines were cultured in medium containing 10% fetal calf serum treated with TRAIL (50 ng/ml) for 6 h, and collected. The arrow shows the location of the PARP cleavage product (85 kDa). The two lanes on the right contain extracts from SkBr3 cells treated with 50 ng of PMA per ml for 20 h to show diminished c-myc expression or left untreated. (B) Cells were treated with TRAIL (50 ng/ml) for 6 h, collected, and analyzed for DNA content by propidium iodide staining. Percentages of cells with sub-G1 DNA content are shown. (C) Immunoblot showing endogenous c-myc and Ran protein levels, using infrared scanning of proteins detected by fluorescently labeled secondary antibodies. FLIP was visualized by chemiluminescence. (D) Quantification of c-myc levels normalized to Ran protein expression for each cell type. The average c-myc level is 0.118% of Ran expression, and the average plus 1 standard deviation is 0.199% of Ran expression. (E and F) TRAIL sensitivity positively correlates with c-myc expression and negatively correlates with FLIP expression. (E) Plot of c-myc level versus percentage of sub-G1 DNA content in TRAIL-sensitive cell lines. (F) Plot of c-myc and FLIP levels in TRAIL-sensitive cell lines.
c-myc overexpression sensitizes TRAIL-resistant cells to TRAIL-mediated cell death.
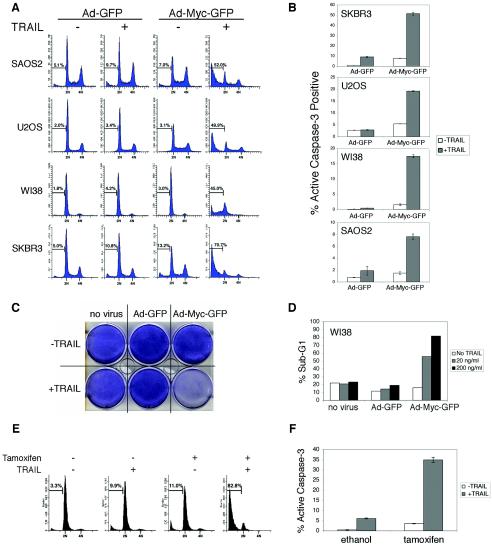
To investigate the pathway of c-myc induced apoptosis in human cells, we used a c-myc- overexpressing adenovirus (Ad-cMyc-GFP) and the GFP-expressing adenovirus Ad-GFP as a control. We used phorbol 12-myristate 13-acetate (PMA)-treated SkBr3 human breast cancer cells as a model to examine the c-myc transcriptional response (7, 46, 47). When exposed to PMA, SkBr3 cells arrest in the G1 phase of the cell cycle and endogenous levels of c-myc decrease rapidly (Fig. 1A) (7, 46, 47). Infecting PMA- treated SkBr3 cells with a c-myc-expressing adenovirus provides a comparison between the c-myc-expressing human cells and those with diminished levels of endogenous c-myc. Addition of TRAIL for 6 h to c-myc-expressing cells resulted in apoptosis in the TRAIL- resistant cells, SAOS2, U2OS, WI-38, and SkBr3 (Fig. 2A and B). Treatment of these cells with TRAIL in the presence of serum or in serum-deprived medium did not result in significant cell death in uninfected cells (data not shown) or in Ad-GFP-infected cells. Since SAOS2 cells do not express p53 and since SkBr3 cells express a mutant p53 deficient in transcription, it appears that c-myc can sensitize cells to TRAIL independently of p53 activation. To confirm that TRAIL mediated a significant apoptotic response, normal human diploid fibroblast WI-38 cells were incubated for 24 h with TRAIL and stained with Coomassie blue (Fig. 2C). Increasing amounts of TRAIL resulted in almost complete cell killing with higher doses of TRAIL (Fig. 2D).
FIG. 2.
c-myc sensitizes TRAIL-resistant cells to TRAIL-induced apoptosis. (A and B) Exogenous c-myc sensitizes TRAIL resistant cells. (A) Serum-deprived WI-38, U2OS, and SAOS2 cells and PMA-treated G1-arrested SkBr3 cells were infected with the indicated adenoviruses (Ad) 24 h prior to TRAIL addition. TRAIL was added to SAOS2 and SkBr3 (100 ng/ml) and WI-38 and U2OS (200 ng/ml) cells for 16 h, collected, fixed, stained for DNA content, and analyzed by flow cytometry. Percentages of cells with sub-G1 DNA content are shown. (B) Serum-deprived cells were infected with the indicated adenoviruses 24 h prior to TRAIL addition (50 ng/ml). At 6 h later, the cells were collected and analyzed for activated caspase 3 by flow cytometry. (C to F) TRAIL kills c-myc-expressing normal human lung fibroblasts. (C) Serum-deprived WI-38 cells were infected with adenovirus, and 24 h later TRAIL (50 ng/ml) was added. After another 24 h, culture medium was removed and the cells were stained with Coomassie blue. (D) Serum-deprived WI-38 cells were infected with adenoviruses (as indicated) for 24 h. The indicated concentrations of TRAIL were added. After 24 h, the cells were collected, fixed, and stained with propidium iodide and the DNA content was analyzed by flow cytometry. (E) myc-ER-expressing WI-38 cells were treated with 500 nM 4-hydroxytamoxifen for 24 h and then with TRAIL (200 ng/ml) for 16 h in the presence of 4-hydroxytamoxifen and analyzed for DNA content as in panel A. (F) myc-ER- expressing WI-38 cells were treated with 500 nM 4-hydroxytamoxifen for 24 h and then with TRAIL for 6 h in the presence of 4-hydroxytamoxifen. The cells were collected and analyzed for activated caspase 3.
In addition, we generated early-passage stably transfected WI-38 cells expressing a myc-ER fusion protein. The myc-ER protein remains localized to the cytoplasm until the addition of 4-hydroxytamoxifen, resulting in the unmasking of the nuclear localization signal of c-myc (38). Within 4 h, nascent mRNA transcripts are synthesized by transactivation of the nuclear myc-ER (data not shown). In agreement with data using c- myc-expressing adenovirus, activation of the myc-ER protein also significantly sensitized these normal cells to TRAIL (Fig. 2E and F).
c-myc decreases FLIP mRNA levels and protein expression.
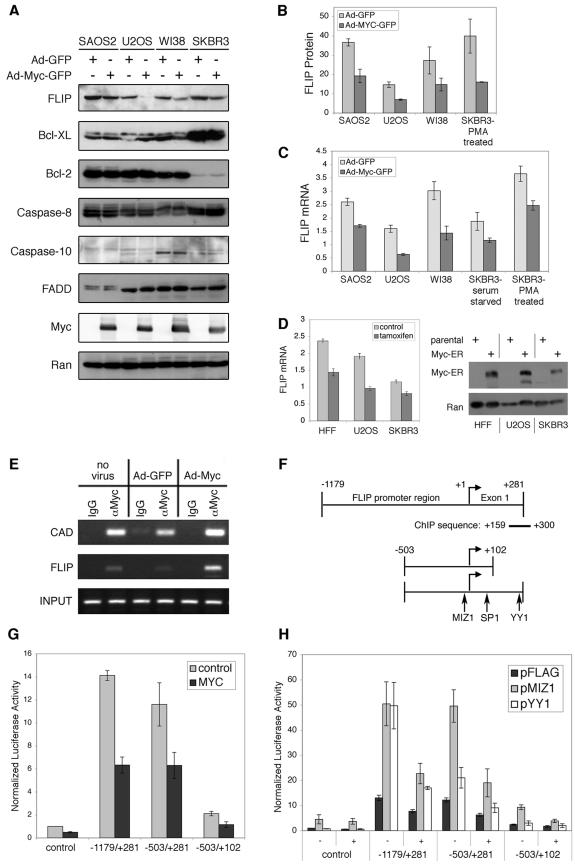
That c-myc can sensitize cells that are otherwise resistant to TRAIL is striking. In an effort to identify a mechanism of c-myc action, we investigated whether c-myc alters the expression of Bcl-XL and the short isoform of FLIP (FLIPS). We examined these two proteins because we had previously identified each of them in a genetic screen as being sufficient to confer resistance to TRAIL in sensitive cells (11), a finding recently confirmed by others (32). Analysis of FLIP, Bcl-XL and Bcl-2, and the other DISC-associated proteins (FADD, caspase 8, and caspase 10) indicated that only the long FLIP isoform was decreased by c-myc expression in all four of the cell lines tested (Fig. 3A). While many FLIP mRNA isoforms are present in most tissue types, only two are expressed at the protein levels, a 55-kDa variant (FLIPL) and a 26-kDa form (FLIPS) (35). FLIPS is minimally or not expressed in SAOS2, U2OS, WI-38, and SkBr3 cells in comparison to FLIPL (data not shown). We quantified FLIP protein levels in c-myc-expressing cells and observed decreases in FLIP expression ranging from 46% in WI-38 cells to 60% in SkBr3 cells (Fig. 3B). We also examined whether c-myc can regulate the TRAIL death receptors DR4 and DR5 and found modest increases in DR5 mRNA in three of the four cell lines examined and a significant increase in DR4 in one cell line (data not shown). If detected, there was no change in TRAIL decoy receptor mRNA levels after c-myc expression (data not shown). None of the proteins that make up the TRAIL DISC, with the exception of FLIP, nor the antiapoptotic Bcl family members Bcl-2 and Bcl-XL, varied in a clear and consistent manner with c-myc expression. Therefore, we focused on determining whether FLIP was a direct target of c-myc repression that mediates myc-induced sensitization to TRAIL.
FIG.3.
c-Myc represses FLIP transcription. (A to D) c-myc expression reduces FLIP protein and mRNA levels. (A) Serum-deprived SAOS2, U2OS, and WI-38 cells and PMA-treated SkBr3 cells were infected with adenoviruses (Ad) (as indicated) for 24 h and then collected, and immunoblotting with antibodies to the indicated proteins was performed. (B) Graph showing quantification of FLIP levels, normalized to Ran, from two separate experiments. (C) Real-time quantitative RT-PCR analysis of FLIP expression was performed using mRNA collected from cells 20 h after adenovirus infection. The results shown were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. (D) (Left panel) Real-time quantitative RT-PCR analysis of FLIP expression, using mRNA collected from serum-starved myc-ER-expressing cells treated with 4-hydroxytamoxifen. At 9 h after 4-hydroxytamoxifen addition, the cells were collected, mRNA was extracted, and quantitative RT-PCR was performed. Results shown were normalized to GAPDH levels. (Right panel) Immunoblot showing myc-ER expression in cells compared with the parental cells. (E to H) c-myc binds the FLIP promoter and represses transcription. (E) Serum-deprived U2OS cells were infected with adenovirus for 24 h, fixed with formaldehyde, and collected. Chromatin immunoprecipitation was performed, followed by PCR amplification of the sequence within the FLIP promoter region shown in panel F. PCR amplification of a c-myc binding region of the CAD gene was used as a positive control. Amplification of the FLIP promoter sequence using 0.5% of the total input is shown. (F) A schematic of the three luciferase reporter plasmids containing the indicated segments of the FLIP promoter is shown. (G and H) U2OS cells were transfected with luciferase reporter constructs containing the putative FLIP promoter sequence shown plus expression plasmids for c-myc, Miz-1, and YY-1 as described in Materials and Methods.
We tested whether c-myc altered FLIP mRNA levels by performing quantitative RT-PCR analysis of the RNA derived from adenovirus-infected cells. We observed significant decreases (35 to 60%) in FLIP mRNA levels in all of the cell lines tested (Fig. 3C). The range of repression of FLIP mRNA by c-myc is similar to the range of repression of FLIP (46 to 60%), suggesting that the regulation of FLIP by c-myc may be predominantly transcriptional. As further confirmation that c-myc represses FLIP transcription, we activated myc in three different myc-ER-expressing cells by using tamoxifen and examined FLIP mRNA levels (Fig. 3D). We observed a c-myc-mediated reduction in FLIP mRNA levels in each of the cell lines tested, including normal HFF.
c-myc binds the FLIP promoter region and represses transcription.
To verify that c-myc can interact with the endogenous FLIP promoter, we performed chromatin immunoprecipitation analysis. We found that c-myc specifically immunoprecipitated the FLIP promoter sequence (Fig. 3E). Activation of c-myc function was confirmed by testing c-myc binding to the E-box-containing promoter region of the CAD gene, a well characterized c-myc target gene (8).
Although more is known about how c-myc activates transcription than about how it can repress, it seems clear is that c-myc does not directly bind to DNA to exert transcriptional repression (65). Instead, it binds to other transcription factors and represses their function. Thus far, Miz-1, SP-1, YY-1, TFII-1, Smad-2, and Smad-3, and NF-Y have all been implicated in c-myc-mediated repression (65). Multiple splice variants of FLIP are found in the National Center for Biotechnology Information GenBank database, and no detailed analysis of the FLIP promoter has been reported. However, an analysis of human chromosome 2q33-q34, which includes the intron-exon organization of FLIP, has been reported by Hadano et al. (23). Of note, the transcriptional start site of the FLIP gene, according to Hadano et al., begins 29 bp downstream of the National Center for Biotechnology Information Reference Sequence for FLIP mRNA (NM_003879). This 29-bp sequence has 18- of 29-bp identity to a sequence 215 bp upstream of the proposed transcriptional start site (23). Therefore, there may be more than one transcriptional initiation site within the human FLIP gene. To include these two potential transcriptional initiation sites in our analysis, we cloned a region encompassing ∼1,200 bp upstream of the first FLIP exon designated in reference 23 and tested it for potential transcriptional regulation by c-myc (Fig. 3F). Examination of this putative FLIP promoter regulatory sequence for known mediators of c-myc repression revealed eight potential Miz-1 binding Inr elements, Py-Py(C)-A-N-(T/A)-Py-Py (12). One potential SP-1 site and one potential YY-1 binding site were also identified within the designated first exon (Fig. 3F). Given the complexity of factors that may interact at the FLIP promoter, we restricted our initial examination to whether c-myc can repress this putative regulatory region. Transfection of a reporter plasmid containing the 1.2-kb putative FLIP regulatory region cloned upstream of the firefly luciferase cDNA in U2OS cells showed a 14-fold increase in luciferase activity compared to the control reporter (Fig. 3G). Coexpressing c-myc reduced luciferase expression by more than 50% (Fig. 3G). These results suggest that this DNA sequence has constitutive basal promoter activity and that c-myc can repress its transcriptional activity. To better delineate the sequences sensitive to c-myc, we removed 680 bp upstream of the transcriptional start site (positions −503 to + 281). We found that the reduction in basal activity was minimal and that this sequence was still sensitive to c-myc (Fig. 3G). Removal of 180 bp from the region downstream of the proposed transcriptional start site (positions −501 + 102) (23) resulted in a significant decrease in activity (Fig. 3G). The YY-1 binding site was removed in this construct, suggesting that it is potentially significant in regulating FLIP transcription. Because the potential SP-1 binding site still remains within this sequence, which has low basal activity, we eliminated it from further analyses. Both Miz-1 and YY-1 activate the FLIP reporter plasmids, and their activity is repressed by c-myc expression (Fig. 3H). Curiously, the reporter plasmid containing bp −503 to +281 of the FLIP promoter was less sensitive to activation by YY-1 than by Miz-1, even though this region contains the putative YY-1 binding site. We conclude from these data that the activation of the FLIP promoter may involve multiple interacting transcription factors, one or more of which is repressed by c-myc.
c-myc expression in vivo represses FLIP.
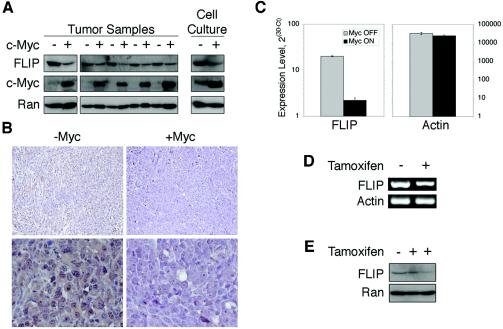
To confirm that our observations were reproducible in vivo, we examined FLIP expression in two different mouse models of myc-induced tumorigenesis. In the first model, we transduced c-myc into Ki-ras-transformed p53−/− murine colonocytes (58) and injected these cells into the flanking region of syngeneic C57BL6/J mice. Tumors grew quickly and were collected 15 days later. Tumors expressing the c-myc transgene had greatly reduced FLIP levels (Figs. 4A and B). By using a second mouse model, we took advantage of the myc-ER system to examine FLIP levels after the conditional activation of myc (69, 70). myc-ER tumors required a continuous supply of 4-hydroxytamoxifen for growth. Nonetheless, in 4-hydroxytamoxifen-deprived animals, small neoplastic lesions persisted and were readily reactivated via readministration of 4-hydroxytamoxifen (D. Yu et al., unpublished data). FLIP levels in the reactivated lesions with functional c-myc were compared to the levels in latent lesions without functional c-myc. FLIP mRNA and protein levels were significantly reduced in tumors with activated c-myc (Fig. 4C to E). These data, taken together, add strong support that c-myc represses FLIP expression in an in vivo setting.
FIG. 4.
c-Myc represses FLIP expression in vivo. (A and B) c-myc-expressing tumor xenografts have diminished FLIP expression. Ki-ras- or Ki-ras/c-myc-transformed p53-null colonocytes were injected subcutaneously into the flanks of mice, and tumors were collected 15 days later. (A) The left panel shows FLIP expression in tumors (with or without c-myc) obtained in two separate experiments. The right panel shows FLIP expression in the cultured cells from one of these experiments prior to injection. (B) Representative sections subjected to immunohistochemical staining for FLIP (brown) from tumors shown in panel A. (C to E) Conditionally activated myc-ER-expressing tumors show reduced FLIP mRNA and protein levels. Myc OFF and “tamoxifen−” refer to small neoplastic lesions from tumor-bearing animals originally treated with but subsequently deprived of 4-hydroxytamoxifen. Myc ON and “tamoxifen +” refer to analogous lesions after restimulation with 4- hydroxytamoxifen. (C) Tumors from these mice were analyzed for FLIP mRNA expression using real-time RT-PCR. Actin levels are shown as controls. (D) Gel electrophoresis of PCR products from real-time RT-PCR analysis depicted in panel C. (E) Immunoblotting of tumor lysates corresponding to samples analyzed in panel C.
c-myc knockdown desensitizes cells to TRAIL.
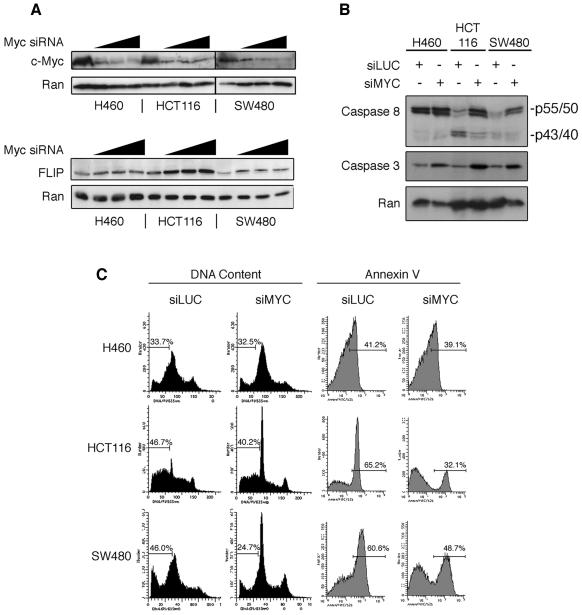
To further test our hypothesis that c-myc controls TRAIL sensitivity, we used siRNA- mediated knockdown of c-myc expression in TRAIL-sensitive cells. Double-stranded siRNA oligonucleotides directed toward c-myc were transfected into H460, HCT116, and SW480 cells, resulting in significant reduction of c-myc protein levels in all three cell lines (Fig. 5A). FLIP levels increased significantly in both HCT116 and SW480 cells but less so in H460 cells. To verify that other myc homologs were not expressed at significant levels in H460 cells, we examined L-myc and N-myc expression by RT-PCR (data not shown). We detected L-myc mRNA expression in the three cell lines, but at much lower levels than c-myc, and there was no detectable N-myc. We have reported previously that cells become more strongly sensitized to TRAIL-induced apoptosis when arrested in G0/G1 using serum deprivation (28). We show (see Fig. 6D) that knockdown of c-myc using siRNA in HCT116 cells (not treated with TRAIL) results in an increase in the G1 phase and decreases in the S and G2 phases of the cell cycle. Therefore, c-myc siRNA increases G0/G1 arrest but decreases TRAIL sensitivity. We interpret these data to mean that the cell cycle effects of c-myc knockdown do not contribute to the decrease in TRAIL-induced apoptosis, but, rather, that the specific knockdown of c-myc levels mediates the desensitization to TRAIL.
FIG. 5.
c-Myc siRNA rescues TRAIL-sensitive cells from TRAIL-induced death. (A) FLIP levels increase in TRAIL-sensitive cells after c-myc mRNA knockdown. Cells were plated and transfected with 50, 100, or 150 nM c-myc siRNA for 36 h. Whole-cell lysates were collected, and immunoblotting for the proteins indicated was performed. (B and C) TRAIL sensitivity is significantly reduced after c-myc knockdown. Cells were transfected with c-myc (siMyc) or luciferase (siLUC) siRNA oligonucleotides for 36 h. (B) TRAIL was added for 6 h, whole-cell lysates were prepared, and immunoblotting was performed. (C) (Left panel) Cells were collected, fixed, and stained with propidium iodide, and the DNA content was measured. Percentages of cells with sub-G1 DNA content are shown. (Right panel) TRAIL was added for 4 h, and the cells were stained with annexin V and analyzed by flow cytometry. Percentages of cells staining positive for annexin V are shown.
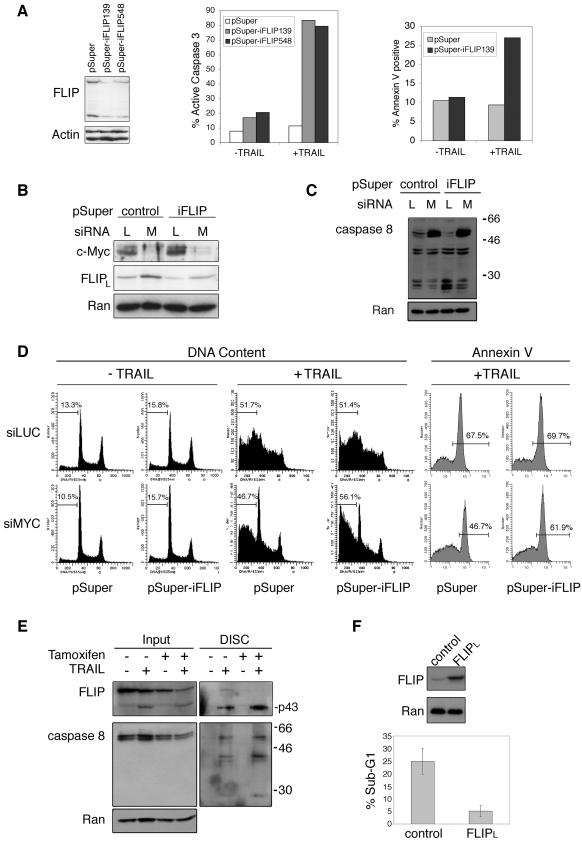
FIG.6.
FLIP is an important mediator of myc-induced sensitization to TRAIL. (A) U2OS cells were transfected with the indicated plasmids. (Left panel) At 48 h later, the cells were collected and immunoblotting was performed. (Center and right panels) Cells were transfected with the indicated plasmids, serum starved 24 h later, and treated with TRAIL for 6 h (50 ng/ml). The cells were collected, stained, and analyzed as shown. (B) HCT116 cells were transfected with the indicated siRNA oligonucleotides and plasmids, collected after 48 h, and analyzed for c-myc and FLIP levels. (C and D) HCT116 cells are resensitized to TRAIL after combined c-myc and FLIP mRNA knockdown. (C) Whole-cell lysates from HCT116 cells transfected and treated with TRAIL for 6 h were collected and analyzed for caspase 8. (D) (Left panel) HCT116 cells were transfected, treated with TRAIL for 6 h, collected, fixed, and stained with propidium iodide. DNA content was measured by flow cytometry, and percentages of cells with sub-G1 DNA content are shown. (Right panel) TRAIL was added for 4 h, and cells were stained with annexin V and analyzed by flow cytometry. Percentages of cells staining positive for annexin V are shown. (E) More cleaved FLIP and caspase 8 is found at TRAIL DISC in cells with activated myc. WI-38 myc-ER cells were serum deprived for 24 h and treated with 4-hydroxytamoxifen for an additional 24 h or left untreated. The cells were collected, TRAIL was added for 30 min, and DISC immunoprecipitation was performed. (F) U2OS cells are made resistant to c-myc action by introduction of exogenous FLIP. (Upper panel) Immunoblot showing the basal expression of FLIP in U2OS cells stably expressing either a control vector or FLIP under the control of a tetracycline-responsive element. This system is leaky, and a low level of exogenous FLIP expression occurs in the absence of tetracycline. (Lower panel) Sub-G1 DNA content of serum-deprived cells infected with c-myc-expressing adenovirus for 24 h and treated with TRAIL (200 ng/ml) for 16 h.
Knocking down c-myc levels and treating the cells with TRAIL significantly reduced the amount of TRAIL-induced death in HCT116 and SW480 cells but left H460 cells unaffected (Fig. 5B and C). As shown in Fig. 5B, c-myc knockdown significantly reduced TRAIL-induced pro-caspase 8 cleavage in HCT116 and SW480 cells. Reduced cleavage of caspase-8 in myc siRNA-treated cells corroborates the flow cytometry results (Fig. 5C) that show less TRAIL-induced death in c-myc siRNA-treated cells. H460 cells underwent less cell death than HCT116 and SW480 cells after treatment with TRAIL under our experimental conditions, and knocking down c-myc expression did not desensitize the H460 cells to TRAIL. These results suggest that there may be different mechanisms of apoptotic activation in the H460 cells than in the HCT116 and SW480 cells. Interestingly, knockdown of c-myc reduced caspase 3 cleavage in the three cell lines (Fig. 5B), suggesting that c-myc may affect other components of the apoptotic machinery that may impact mitochondrial cytochrome c release. It appears that this change in caspase 3 cleavage is not functionally significant in H460 cells, however, since there were no observed differences in TRAIL-induced cell death with or without c-myc knockdown (Fig. 5C). While it is not entirely clear why c-myc knockdown does not affect TRAIL-induced death in H460 cells, we propose that the TRAIL DISC is not fully engaged in H460 cells, as seen by lack of caspase 8 cleavage in Fig. 5B, while it is fully engaged in HCT116 and SW480 cells. Therefore, the slight increase in FLIP expression by c-myc knockdown in H460 cells is apparently not functionally significant since the levels of TRAIL-induced cell death with or without c-myc siRNA do not change. The change in FLIP expression in HCT116 and SW480 cells after c-myc knockdown does appear to be functionally significant, however, because TRAIL-induced cleavage of caspase 8 is blocked after c-myc knockdown, suggesting that the TRAIL DISC is not fully engaged.
FLIP knockdown resensitizes cells to TRAIL.
Examination of the relationship between c-myc and FLIP levels in TRAIL-sensitive cells (Fig. 1F) shows an inverse correlation between c-myc and FLIP (correlation coefficient = −0.782). A similar inverse correlation between FLIP and TRAIL sensitivity in the TRAIL-sensitive cells also exists (correlation coefficient = −0.783). These results suggest that an important relationship exists between c-myc expression, FLIP expression, and TRAIL sensitivity in the panel of TRAIL-sensitive cell lines used in these studies. To directly test the functional importance of FLIP repression by myc, we examined whether knocking down FLIP in c-myc siRNA-treated cells might resensitize these cells to TRAIL. Using plasmid vector-mediated expression of short-hairpin RNA sequences that would be processed in vivo to siRNA oligonucleotides, we identified two sequences that each elicit significant down-regulation of FLIP protein levels and sensitization to TRAIL (Fig. 6A). We verified the combined FLIP and c-myc knockdown in HCT116 cells (Fig. 6B) and observed significant knockdown of c-myc protein and a modest but detectable knockdown of FLIP. Although knocking down FLIP in c-myc siRNA-treated cells resulted in FLIP protein levels that were similar to those in control cells without knockdown (Fig. 6B, compare the first and fourth lanes), the changes in caspase 8 cleavage and the amount of cell death were quite different (Fig. 6C and D). myc knockdown results in significant protection from TRAIL-induced pro-caspase 8 cleavage, with or without FLIP knockdown. However, the combined knockdown of c-myc and FLIP resulted in an increase in sub-G1 DNA content and annexin V staining, compared to the effects of knocking down c-myc alone (Fig. 6D), and in the amount of cleaved caspase 8 (Fig. 6C). We interpret these results to mean that c-myc repression of FLIP expression plays a significant, but not a singular, role in the ability of c-myc to sensitize cells to TRAIL.
In an attempt to better determine the functional importance of FLIP repression by c-myc in TRAIL sensitization, we examined whether FLIP levels were affected by c-myc at the TRAIL-induced DISC in WI-38 myc-ER cells (Fig. 6E). Cells with activated c-myc had lower levels of FLIP protein. The TRAIL-induced DISC of tamoxifen-treated cells had no detectable uncleaved FLIP, unlike the DISC of cells not treated with tamoxifen. Also, cells treated with tamoxifen had more cleaved FLIP at the DISC than did cells not treated with tamoxifen. We also found more pro-caspase 8 and cleaved caspase 8 at the DISC of cells with activated c-myc. We interpret these results to mean that c-myc activation enhances DISC activation, resulting in more caspase 8 and FLIP cleavage. It appears that cleaved FLIP at the DISC of c-myc-expressing cells, under these conditions, does not inhibit caspase-8 cleavage. These results agree with the emerging paradigm that, unlike the short form of FLIP, the processed form of FLIPL is not a universal inhibitor of caspase 8 activation (36). Instead, it has been shown recently that low levels of FLIP can activate caspase 8 at the DISC while high levels of FLIP block its activation (13, 45). Therefore, c-myc repression of FLIP expression reduces the amount of FLIP available sufficiently to block caspase 8 activation, resulting in increased amounts of cleaved caspase 8 and cleaved FLIP at the DISC. It is also possible that low FLIP levels in cells with activated c-myc may act to enhance caspase 8 activation at the TRAIL DISC.
As another method to examine the importance of FLIP repression by myc in the TRAIL response, we generated stable U20S cells that express FLIP under a tetracycline-driven promoter. We found the basal expression of FLIP in these cells to be slightly higher than in control cells and used these cells without the addition of tetracycline for our experiment. We compared the effects of TRAIL after treatment with c-myc-expressing adenovirus and found that cells expressing exogenous FLIP became resistant to c-myc-induced TRAIL sensitization (Fig. 6F). This experiment adds support to our hypothesis that c-myc can sensitize cells to TRAIL by specifically repressing FLIP expression.
We conclude from these combined results that FLIP levels play a critical role in activation of the TRAIL DISC. High FLIP levels overcame c-myc-induced TRAIL sensitization (Fig. 6F). Repressing FLIP expression in myc-ER-activated cells resulted in increased caspase 8 and FLIP cleavage at the DISC (Fig. 6E). Reducing FLIP levels in c-myc siRNA-treated cells increased their sensitization to TRAIL but did not recapitulate the TRAIL-induced death phenotype if c-myc levels were elevated (Fig. 6B to D). Therefore, it appears that c-myc may affect other components of the TRAIL signaling pathway, e.g., DR4 or DR5, that may contribute to the sensitization to TRAIL seen in high-c-myc-expressing cells. In our analysis, however, FLIP is the only DISC member that we identified to be universally and directly regulated by c-myc.
DISCUSSION
TRAIL holds great potential as an anticancer agent. It can selectively kill cancer cells while leaving most normal cells unharmed. However, not all cancer cells are sensitive to TRAIL. An interesting hypothesis that emerges from the present studies is that high c-myc levels may be an Achilles' heel of cancer cells regarding susceptibility to TRAIL. It is well established that FLIP expression is an important variable determining TRAIL sensitivity (11, 33, 63). In the data we present here, we have shown that c-myc can repress FLIP transcription and thereby promote TRAIL sensitivity. While this direct connection has not been investigated before, previous studies have hinted at a connection between c-myc expression, FLIP levels, and TRAIL sensitivity.
For example, an association between c-myc expression, TRAIL sensitivity, and FLIP levels in studies using Burkitt's lymphoma (BL) cell lines has emerged. All BL tumors share the translocation of Ig and MYC genes (reviewed in reference 42). BL is associated with Epstein-Barr virus (EBV) infection, although the pathogenetic mechanisms leading to c-myc translocation remain less well understood. A recent study examined BL cell lines for TRAIL sensitivity and found a strong correlation with negative EBV status and sensitivity to TRAIL (48). Three of the EBV-positive TRAIL-resistant cell lines described in reference 48 were found to have high FLIP mRNA and protein levels (61). While neither of these studies examined the BL lines for c-myc expression, other studies found, using matched pairs of EBV-positive and -negative BL cells, that EBV-positive BL cells have significantly decreased c-myc levels (6, 57). Therefore, an association exists in BL cells, correlating reduced TRAIL sensitivity with positive EBV status, low c-myc expression, and high FLIP levels.
Also in agreement with our findings, N-myc, the c-myc homologue found overexpressed in neuroblastomas, while not shown to sensitize resistant cells, was found to increase the apoptotic response to a high dose of TRAIL (39). Another recent report showed that c-myc expression accelerated interleukin-6 (IL-6)-mediated down-regulation of FLIP (1). While the mechanism by which c-myc collaborates with IL-6 to down-regulate c-FLIP was not elucidated, our results provide a mechanism for c-myc-mediated FLIP repression that works in concert with IL-6. In a recent study examining the basal levels of DISC components in lung cancer cell lines, it was shown that small cell lung cancer cells with myc amplification were more resistant to TRAIL, had reduced expression of proapoptotic DISC components, and showed increased expression of FLIP (59). These findings, which may seem to contradict our conclusions, agree with our results using c-myc siRNA knockdown in H460 cells, a cancer cell line derived from the pleural effusion of a large cell lung carcinoma, and suggest that c-myc may not regulate TRAIL sensitivity in lung cancer. While we observed only a modest change in FLIP expression in H460 cells by c-myc siRNA, we did observe both dramatic protection by c-myc knockdown against TRAIL-induced death in colon cancer cells and a coincident increase in FLIP expression (Fig. 5). In support of our findings, c-myc was recently found in a screen using a siRNA library to identify modulators of TRAIL-induced apoptosis in HeLa cervical adenocarcinoma cells (5). The authors of that study did not explore why c-myc knockdown might sensitize cells to TRAIL but instead focused on c-myc regulation by the Wnt signaling pathway and found that siRNA to Wnt transducers TCF4 and DVL2 also regulated TRAIL sensitivity.
We examined serial analysis of gene expression (SAGE) data collected by the Cancer Genome Anatomy Project of the National Cancer Institute (available at http://cgap.nci.nih.gov/Genes/GeneFinder) for c-myc and FLIP expression in cDNA libraries constructed from normal and cancerous tissues. SAGE data were present for both c-myc and FLIP in 59 different cDNA libraries. We extracted expression values from these 59 cDNA libraries and found that 33 libraries (56%) had more c-myc expression than FLIP expression, 12 libraries (20%) had more FLIP expression than c-myc expression, and 14 libraries (24%) had equivalent c- myc and FLIP expression. Therefore, a total of 45 of 59 libraries (76%) had an inverse relationship between c-myc and FLIP expression. This trend is in agreement with the correlation we observed between c-myc and FLIP expression in the TRAIL-sensitive cells in our cell panel (Fig. 1F). While a more comprehensive analysis of tissue types is necessary to confirm whether there is a universal inverse relationship between c-myc and FLIP expression that spans tissue types, the comparison of c-myc and FLIP expression by SAGE analysis gives in vivo significance to the potential for endogenous human c-myc to negatively regulate FLIP expression.
It is known that c-myc can induce apoptosis in cells deprived of survival factors (21). Since this key observation was made, an emerging view to explain why an oncogene would induce apoptosis is that a cell's proliferative and apoptotic pathways are coupled and that apoptosis is suppressed as long as pro-survival signaling pathways are intact and activated (54). Much work has been done to identify the genes regulated by c-myc, including SAGE gene expression analyses and genome-wide c-myc E-box binding studies (22, 44, 71), but it is still not clear which c-myc targets are essential for the activation of apoptosis. For example, we found the proapoptotic Bcl family member Bax to be a transcriptional target of c-myc (47), but this induction is not observed in all cell systems (31, 60). However, recent observations using Bax−/− cells have concluded that Bax loss impairs c-myc-induced apoptosis (17, 31) and that c-myc binds to the Bax locus as documented by chromatin immunoprecipitation analysis (22, 71). Repression of the prosurvival Bcl family members Bcl-2 and Bcl-XL by c-myc has also been reported (16, 18, 41), but we show here that there is not a similar pattern of regulation by c-myc in all the cell lines we tested (Fig. 3A). While exact mechanisms to explain c-myc-induced alterations of proteins involved in maintaining mitochondrial membrane integrity may not be consistent across cell and tissue types, c-myc does exert a powerful effect on the mitochondria (30, 31, 47, 60).
We observed a direct linear correlation between c-myc levels and TRAIL sensitivity (Fig. 1E). We also observed an inverse linear relationship between c-myc and FLIP in the TRAIL-sensitive cells (Fig. 1F). We propose that c-myc regulates FLIP expression directly and that c-myc repression of FLIP contributes to TRAIL sensitivity. FLIP is an important regulator of death receptor-mediated apoptosis. Up-regulation of FLIP was detected in many tumors (9, 27, 62), and expression of FLIP in transgenic mice results in escape from T-cell immune surveillance and subsequent tumor growth (15, 43). FLIP inhibits death receptor-triggered apoptosis through its recruitment to the DISC and inhibition of caspase 8 cleavage. We observed that c-myc knockdown results in increased FLIP expression, reduced caspase 8 cleavage, and reduced sensitivity to TRAIL in HCT116 and SW480 cells. We also observed that reducing FLIP expression back to control levels in HCT116 cells (Fig. 6B) did not recapitulate the TRAIL response seen in the control cells (Fig. 6C and D). However, we observed more full-length FLIP at the TRAIL DISC in cells without activated c-myc and more caspase 8 at the TRAIL DISC of cells with activated c-myc (Fig. 6E), suggesting that c-myc enhances activation of the DISC. Because there is less FLIP in cells with activated c-myc and there is no detectable full-length FLIP at the DISC of these cells, we infer that c-myc repression of FLIP is sufficient to permit the efficient activation of the DISC. Furthermore, increased FLIP expression in TRAIL-resistant U2OS cells protects them from myc-induced sensitization to TRAIL (Fig. 6F). We interpret these results to mean that FLIP is a significant target regulated by c-myc, but not necessarily a singular one, that contributes to the ability of c-myc to sensitize cells to TRAIL.
We propose that elevated c-myc expression in human cancers offer an opportunity for targeted destruction by TRAIL. Elevated or deregulated expression of c-myc has been identified in a wide range of human cancers including cancers of the breast, colon, and cervix, osteosarcomas, glioblastomas, melanomas, lymphomas, and leukemias (49). Oncogenically activated c-myc is also often associated with aggressive, poorly differentiated tumors (49). The observations shown here place the mechanism of c-myc sensitization to TRAIL upstream of the mitochondria. Therefore, elevated c-myc expression in cancer cells resistant to mitochondrial membrane destabilization, for example because of Bcl-2/Bcl-XL overexpression, may confer TRAIL sensitivity by repressing FLIP levels. In addition, p53 function is not an essential mediator of c-myc sensitization to TRAIL. We observed that FLIP expression was down-regulated consistently by c-myc in cell lines having wild-type, mutant, or deleted p53. Therefore, c-myc overexpression in tumors should still confer sensitivity to TRAIL irrespective of the p53 status. We suspect that a number of changes in human tumors that affect apoptotic signaling (e.g., FLIP, Bcl-2, Bcl-XL, and IAP family member) may influence the ability of a cell to undergo apoptosis in response to TRAIL. We do not propose here that FLIP is the only factor relevant to c-myc-induced sensitization to TRAIL, but it appears to be a significant one. In summary, c-myc plays an important role in determining cellular sensitivity to TRAIL. We propose the selective targeting of c-myc-overexpressing cancers by taking advantage of the ability of c-myc to repress FLIP transcription and sensitize cells to TRAIL.
Acknowledgments
We thank Martin Eilers for the Miz-1 plasmid; Michael Goldschmidt for his help with the analysis of immunohistochemical data; and Timothy F. Burns, Seok-Hyun Kim, Kimberly A. Scata, E. Robert McDonald III, Paul G. Corn, and other members of the El-Deiry laboratory for many helpful discussions.
This work was supported by funds from the Howard Hughes Medical Institute (W.S.E.-D.), NIH grant (CA102709 (A.T.-T.), and NIH grants CA75138, CA098101, and CA105008 (W.S.E.-D.). W.S.E.-D. is an Assistant Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Amanullah, A., D. A. Liebermann, and B. Hoffman. 2002. Deregulated c-Myc prematurely recruits both type I and II CD95/Fas apoptotic pathways associated with terminal myeloid differentiation. Oncogene 21:1600-1610. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi, A. 2002. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat. Rev. Cancer 2:420-430. [DOI] [PubMed] [Google Scholar]
- 3.Ashkenazi, A., and V. M. Dixit. 1998. Death receptors: signaling and modulation. Science 281:1305-1308. [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi, A., R. C. Pai, S. Fong, S. Leung, D. A. Lawrence, S. A. Marsters, C. Blackie, L. Chang, A. E. McMurtrey, A. Hebert, L. DeForge, I. L. Koumenis, D. Lewis, L. Harris, J. Bussiere, H. Koeppen, Z. Shahrokh, and R. H. Schwall. 1999. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Investig. 104:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aza-Blanc, P., C. L. Cooper, K. Wagner, S. Batalov, Q. L. Deveraux, and M. P. Cooke. 2003. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12:627-637. [DOI] [PubMed] [Google Scholar]
- 6.Baran-Marszak, F., R. Fagard, B. Girard, S. Camilleri-Broet, F. Zeng, G. M. Lenoir, M. Raphael, and J. Feuillard. 2002. Gene array identification of Epstein Barr virus-regulated cellular genes in EBV-converted Burkitt lymphoma cell lines. Lab. Investig. 82:1463-1479. [DOI] [PubMed] [Google Scholar]
- 7.Blagosklonny, M. V., N. S. Prabhu, and W. S. El-Deiry. 1997. Defects in p21 WAF1/CIP1, Rb, and c-myc signaling in phorbol ester-resistant cancer cells. Cancer Res. 57:320-325. [PubMed] [Google Scholar]
- 8.Boyd, K. E., and P. J. Farnham. 1997. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 17:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullani, R. R., B. Huard, I. Viard-Leveugle, H. R. Byers, M. Irmler, J. H. Saurat, J. Tschopp, and L. E. French. 2001. Selective expression of FLIP in malignant melanocytic skin lesions. J. Investig. Dermatol. 117:360-364. [DOI] [PubMed] [Google Scholar]
- 10.Burns, T., E. Bernhard, and W. El-Diery. 2001. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 20:4601-4612. [DOI] [PubMed] [Google Scholar]
- 11.Burns, T. F., and W. S. El-Deiry. 2001. Identification of inhibitors of TRAIL- induced death (ITIDs) in the TRAIL-sensitive colon carcinoma cell line SW480 using a genetic approach. J. Biol. Chem. 276:37879-37886. [DOI] [PubMed] [Google Scholar]
- 12.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 13.Chang, D. W., Z. Xing, Y. Pan, A. Algeciras-Schimnich, B. C. Barnhart, S. Yaish-Ohad, M. E. Peter, and X. Yang. 2002. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 21:3704-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danielian, P. S., R. White, S. A. Hoare, S. E. Fawell, and M. G. Parker. 1993. Identification of residues in the estrogen receptor that confer differential sensitivity to estrogen and hydroxytamoxifen. Mol. Endocrinol. 7:232-240. [DOI] [PubMed] [Google Scholar]
- 15.Djerbi, M., V. Screpanti, A. I. Catrina, B. Bogen, P. Biberfeld, and A. Grandien. 1999. The inhibitor of death receptor signaling, FLICE-inhibitory protein defines a new class of tumor progression factors. J. Exp. Med. 190:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eischen, C. M., G. Packham, J. Nip, B. E. Fee, S. W. Hiebert, G. P. Zambetti, and J. L. Cleveland. 2001. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene 20:6983-6993. [DOI] [PubMed] [Google Scholar]
- 17.Eischen, C. M., M. F. Roussel, S. J. Korsmeyer, and J. L. Cleveland. 2001. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol. Cell. Biol. 21:7653-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eischen, C. M., D. Woo, M. F. Roussel, and J. L. Cleveland. 2001. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol. Cell. Biol. 21:5063-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 20.El-Deiry, W. S. 2001. Insights into cancer therapeutic design based on p53 and TRAIL receptor signaling. Cell Death Differ 8:1066-1075. [DOI] [PubMed] [Google Scholar]
- 21.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadano, S., Y. Yanagisawa, J. Skaug, K. Fichter, J. Nasir, D. Martindale, B. F. Koop, S. W. Scherer, D. W. Nicholson, G. A. Rouleau, J. Ikeda, and M. R. Hayden. 2001. Cloning and characterization of three novel genes, ALS2CR1, ALS2CR2, and ALS2CR3, in the juvenile amyotrophic lateral sclerosis (ALS2) critical region at chromosome 2q33-q34: candidate genes for ALS2. Genomics 71:200-213. [DOI] [PubMed] [Google Scholar]
- 24.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hueber, A. O., M. Zornig, D. Lyon, T. Suda, S. Nagata, and G. I. Evan. 1997. Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science 278:1305-1309. [DOI] [PubMed] [Google Scholar]
- 26.Ichikawa, K., W. Liu, L. Zhao, Z. Wang, D. Liu, T. Ohtsuka, H. Zhang, J. D. Mountz, W. J. Koopman, R. P. Kimberly, and T. Zhou. 2001. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat. Med. 7:954-960. [DOI] [PubMed] [Google Scholar]
- 27.Irmler, M., M. Thome, M. Hahne, P. Schneider, K. Hofmann, V. Steiner, J. L. Bodmer, M. Schroter, K. Burns, C. Mattmann, D. Rimoldi, L. E. French, and J. Tschopp. 1997. Inhibition of death receptor signals by cellular FLIP. Nature 388:190-195. [DOI] [PubMed] [Google Scholar]
- 28.Jin, Z., D. T. Dicker, and W. S. El-Deiry. 2002. Enhanced sensitivity of G1 arrested human cancer cells suggests a novel therapeutic strategy using a combination of simvastatin and TRAIL. Cell Cycle 1:82-89. [PubMed] [Google Scholar]
- 29.Jo, M., T. H. Kim, D. W. Seol, J. E. Esplen, K. Dorko, T. R. Billiar, and S. C. Strom. 2000. Apoptosis induced in normal human hepatocytes by tumor necrosis factor-related apoptosis-inducing ligand. Nat. Med. 6:564-567. [DOI] [PubMed] [Google Scholar]
- 30.Juin, P., A. O. Hueber, T. Littlewood, and G. Evan. 1999. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 13:1367-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juin, P., A. Hunt, T. Littlewood, B. Griffiths, L. B. Swigart, S. Korsmeyer, and G. Evan. 2002. c-Myc functionally cooperates with Bax to induce apoptosis. Mol. Cell. Biol. 22:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, I. K., Y. K. Jung, D. Y. Noh, Y. S. Song, C. H. Choi, B. H. Oh, and E. S. Masuda. 2003. Functional screening of genes suppressing TRAIL-induced apoptosis: distinct inhibitory activities of Bcl-XL and Bcl-2. Br. J. Cancer 88:910-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, K., M. J. Fisher, S.-Q. Xu, and W. S. El-Deiry. 2000. Molecular determinants of response to TRAIL in killing of normal and cancer cells. Clin. Cancer Res. 6:335-346. [PubMed] [Google Scholar]
- 34.Klefstrom, J., I. Vastrik, E. Saksela, J. Valle, M. Eilers, and K. Alitalo. 1994. c-Myc induces cellular susceptibility to the cytotoxic action of TNF-alpha. EMBO J. 13:5442-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krueger, A., S. Baumann, P. H. Krammer, and S. Kirchhoff. 2001. FLICE- inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol. Cell. Biol. 21:8247-8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krueger, A., I. Schmitz, S. Baumann, P. H. Krammer, and S. Kirchhoff. 2001. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 276:20633-20640. [DOI] [PubMed] [Google Scholar]
- 37.Lawrence, D., Z. Shahrokh, S. Marsters, K. Achilles, D. Shih, B. Mounho, K. Hillan, K. Totpal, L. DeForge, P. Schow, J. Hooley, S. Sherwood, R. Pai, S. Leung, L. Khan, B. Gliniak, J. Bussiere, C. A. Smith, S. S. Strom, S. Kelley, J. A. Fox, D. Thomas, and A. Ashkenazi. 2001. Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat. Med. 7:383-385. [DOI] [PubMed] [Google Scholar]
- 38.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutz, W., S. Fulda, I. Jeremias, K. M. Debatin, and M. Schwab. 1998. MycN and IFNgamma cooperate in apoptosis of human neuroblastoma cells. Oncogene 17:339-346. [DOI] [PubMed] [Google Scholar]
- 40.MacLachlan, T. K., and W. S. El-Deiry. 2003. Identification of DNA-binding of tumor suppressor genes by chromatin immunoprecipitation. Methods Mol. Biol. 223:129-133. [DOI] [PubMed] [Google Scholar]
- 41.Maclean, K. H., U. B. Keller, C. Rodriguez-Galindo, J. A. Nilsson, and J. L. Cleveland. 2003. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol. Cell. Biol. 23:7256-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magrath, I. 1990. The pathogenesis of Burkitt's lymphoma. Adv. Cancer Res. 55:133-270. [DOI] [PubMed] [Google Scholar]
- 43.Medema, J. P., J. de Jong, T. van Hall, C. J. Melief, and R. Offringa. 1999. Immune escape of tumors in vivo by expression of cellular FLICE-inhibitory protein. J. Exp. Med. 190:1033-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC- regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Micheau, O., M. Thome, P. Schneider, N. Holler, J. Tschopp, D. W. Nicholson, C. Briand, and M. G. Grutter. 2002. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 277:45162-45171. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell, K. O., and W. S. El-Deiry. 1999. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 10:223-230. [PubMed] [Google Scholar]
- 47.Mitchell, K. O., M. S. Ricci, T. Miyashita, D. T. Dicker, Z. Jin, J. C. Reed, and W. S. El-Deiry. 2000. Bax is a transcriptional target and mediator of c-myc- induced apoptosis. Cancer Res. 60:6318-6325. [PubMed] [Google Scholar]
- 48.Mouzakiti, A., and G. Packham. 2003. Regulation of tumour necrosis factor- related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Burkitt's lymphoma cell lines. Br. J. Haematol. 122:61-69. [DOI] [PubMed] [Google Scholar]
- 49.Nesbit, C. E., J. M. Tersak, and E. V. Prochownik. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18:3004-3016. [DOI] [PubMed] [Google Scholar]
- 50.Ngo, C. V., M. Gee, N. Akhtar, D. Yu, O. Volpert, R. Auerbach, and A. Thomas-Tikhonenko. 2000. An in vivo function for the transforming Myc protein: elicitation of the angiogenic phenotype. Cell Growth Differ. 11:201-210. [PMC free article] [PubMed] [Google Scholar]
- 51.Ozoren, N., M. J. Fisher, K. Kim, C. X. Liu, A. Genin, Y. Shifman, D. T. Dicker, N. B. Spinner, N. A. Lisitsyn, and W. S. El-Deiry. 2000. Homozygous deletion of the death receptor DR4 gene in a nasopharyngeal cancer cell line is associated with TRAIL resistance. Int. J. Oncol. 16:917-925. [DOI] [PubMed] [Google Scholar]
- 52.Ozoren, N., K. Kim, T. F. Burns, D. T. Dicker, A. D. Moscioni, and W. S. El- Deiry. 2000. The caspase 9 inhibitor Z-LEHD-FMK protects human liver cells while permitting death of cancer cells exposed to tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 60:6259-6265. [PubMed] [Google Scholar]
- 53.Pear, W. S., J. P. Miller, L. Xu, J. C. Pui, B. Soffer, R. C. Quackenbush, A. M. Pendergast, R. Bronson, J. C. Aster, M. L. Scott, and D. Baltimore. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92:3780-3792. [PubMed] [Google Scholar]
- 54.Pelengaris, S., M. Khan, and G. Evan. 2002. c-myc: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 55.Prabhu, N. S., M. V. Blagosklonny, Y:-U. Zeng, G. S. Wu, T. Waldman, and W. S. El-Deiry. 1996. Suppression of cancer cell growth by adenovirus expressing p21WAF1/CIP1 deficient in PCNA interaction. Clin. Cancer Res. 2:1221-1229. [PubMed] [Google Scholar]
- 56.Roth, W., S. Isenmann, U. Naumann, S. Kugler, M. Bahr, J. Dichgans, A. Ashkenazi, and M. Weller. 1999. Locoregional Apo2L/TRAIL eradicates intracranial human malignant glioma xenografts in athymic mice in the absence of neurotoxicity. Biochem. Biophys. Res. Commun. 265:479-483. [DOI] [PubMed] [Google Scholar]
- 57.Ruf, I. K., P. W. Rhyne, H. Yang, C. M. Borza, L. M. Hutt-Fletcher, J. L. Cleveland, and J. T. Sample. 1999. Epstein-Barr virus regulates c-Myc, apoptosis, and tumorigenicity in Burkitt lymphoma. Mol. Cell. Biol. 19:1651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sevignani, C., P. Wlodarski, J. Kirillova, W. E. Mercer, K. G. Danielson, R. V. Iozzo, and B. Calabretta. 1998. Tumorigenic conversion of p53-deficient colon epithelial cells by an activated Ki-ras gene. J. Clin. Investig. 101:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shivapurkar, N., J. Reddy, H. Matta, U. G. Sathyanarayana, C. X. Huang, S. Toyooka, J. D. Minna, P. M. Chaudhary, and A. F. Gazdar. 2002. Loss of expression of death-inducing signaling complex (DISC) components in lung cancer cell lines and the influence of MYC amplification. Oncogene 21:8510-8514. [DOI] [PubMed] [Google Scholar]
- 60.Soucie, E. L., M. G. Annis, J. Sedivy, J. Filmus, B. Leber, D. W. Andrews, and L. Z. Penn. 2001. Myc potentiates apoptosis by stimulating Bax activity at the mitochondria. Mol. Cell. Biol. 21:4725-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tepper, C. G., and M. F. Seldin. 1999. Modulation of caspase-8 and FLICE- inhibitory protein expression as a potential mechanism of Epstein-Barr virus tumorigenesis in Burkitt's lymphoma. Blood 94:1727-1737. [PubMed] [Google Scholar]
- 62.Thomas, R. K., A. Kallenborn, C. Wickenhauser, J. L. Schultze, A. Draube, M. Vockerodt, D. Re, V. Diehl, and J. Wolf. 2002. Constitutive expression of c- FLIP in Hodgkin and Reed-Sternberg cells. Am. J. Pathol. 160:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 64.Walczak, H., R. E. Miller, K. Ariail, B. Gliniak, T. S. Griffith, M. Kubin, W. Chin, J. Jones, A. Woodward, T. Le, C. Smith, P. Smolak, R. G. Goodwin, C. T. Rauch, J. C. Schuh, and D. H. Lynch. 1999. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5:157-163. [DOI] [PubMed] [Google Scholar]
- 65.Wanzel, M., S. Herold, and M. Eilers. 2003. Transcriptional repression by Myc. Trends Cell Biol. 13:146-150. [DOI] [PubMed] [Google Scholar]
- 66.Woods, D., D. Parry, H. Cherwinski, E. Bosch, E. Lees, and M. McMahon. 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 17:5598-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, G. S., and W. S. El-Deiry. 1996. Apoptotic death of tumor cells correlates with chemosensitivity, independent of p53 or bcl-2. Clin Cancer Res. 2:623-633. [PubMed] [Google Scholar]
- 68.Xu, D., N. Popov, M. Hou, Q. Wang, M. Bjorkholm, A. Gruber, A. R. Menkel, and M. Henriksson. 2001. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 98:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, D., D. Allman, M. H. Goldschmidt, M. L. Atchison, J. G. Monroe, and A. Thomas-Tikhonenko. 2003. Oscillation between B-lymphoid and myeloid lineages in Myc-induced hematopoietic tumors following spontaneous silencing/reactivation of the EBF/Pax5 pathway. Blood 101:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu, D., and A. Thomas-Tikhonenko. 2002. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene 21:1922-1927. [DOI] [PubMed] [Google Scholar]
- 71.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]