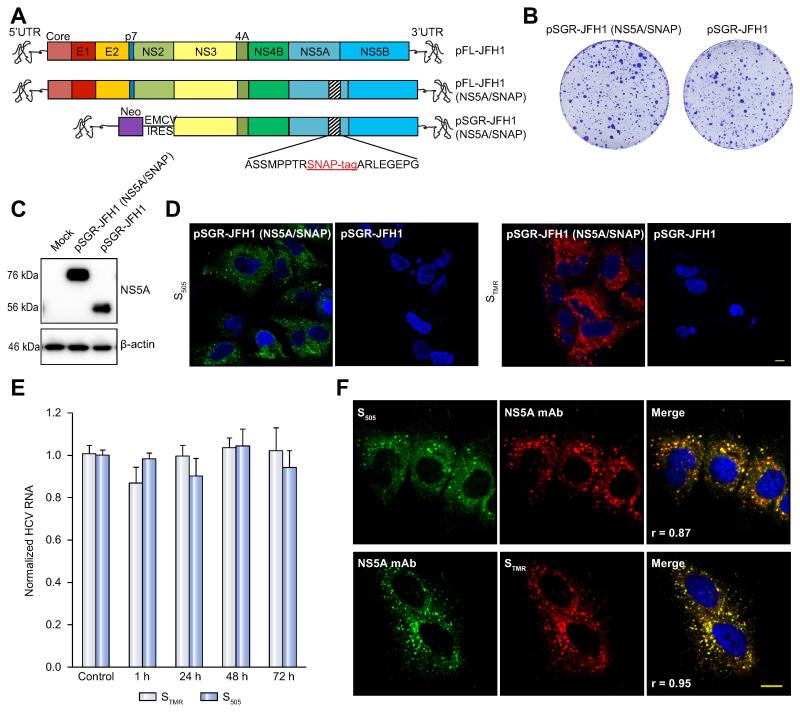
Figure 1. Development and characterization of SNAP tagged HCV genomes for live cell pulse-chase imaging.
(A). Schematic of viral constructs. A SNAP tag was inserted into a known tolerated insertion site within domain III of NS5A in the JFH1 strain to create plasmids encoding full-length FL-JFH1(NS5A/SNAP) and the subgenomic replicon SGR-JFH1(NS5A/SNAP).
(B). Colony formation assay of JFH-1 subgenomic replicons in Huh7.5.1 cells. Huh7.5.1 cells were transfected with in vitro transcribed RNA encoding the indicated constructs and G418-resistant colonies were visualized by Crystal Violet staining 21 days post-transfection.
(C). Immunoblot analysis of NS5A protein expression from cell lysates prepared from the indicated subgenomic replicon cells. β-actin is shown as a loading control.
(D). Living cells expressing the indicated subgenomic replicons with or without the SNAP tag were labeled with 5μM green fluorescent SNAP-Cell 505 (S505) or 3μM red fluorescent SNAP-Cell TMR-Star (STMR) for 15min. Nuclei were counterstained with DAPI. Scale bar, 10μm.
(E). SGR-JFH1(NS5A/SNAP) subgenomic replicon cells were either mock treated or stained with the indicated labeling reagent as in panel D and then incubated in fresh medium for the indicated times. HCV RNA was quantitated by qRT-PCR. Values are means ± SEM of three independent experiments and normalized to control.
(F). Specificity of SNAP labeling for NS5A protein. SGR-JFH1(NS5A/SNAP) subgenomic replicon cells were labeled with either STMR or S505 as above before fixing and immunostaining for NS5A. Scale bar, 10 μm.