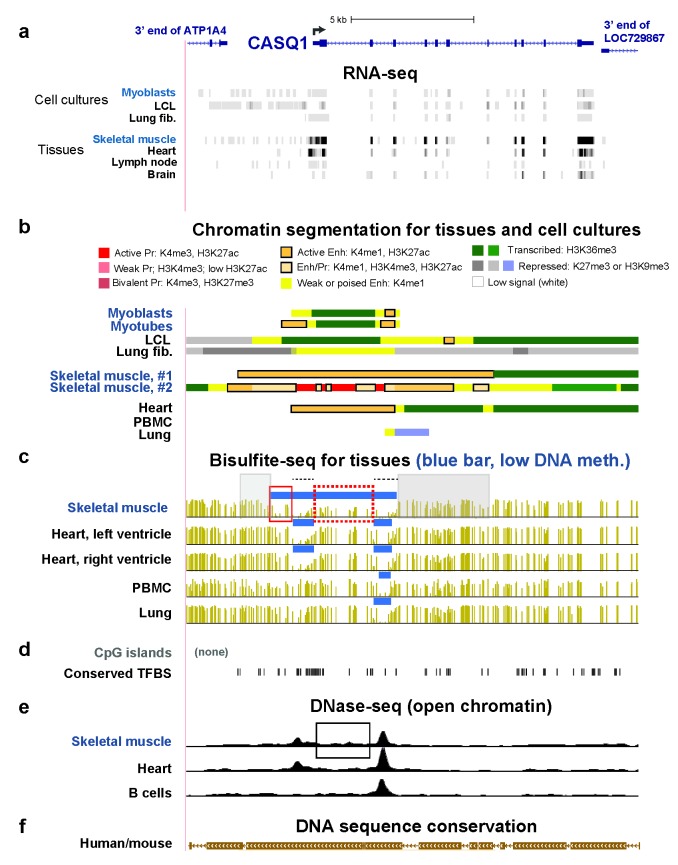
Figure 2.
CASQ1 SkM-specific and SkM- and heart-specific enhancer chromatin overlaps regions of SkM-only DNA hypomethylation. (a) RefSeq gene depictions and RNA-seq data for the CASQ1 gene region and the ends of the neighboring genes at chr1:160,155,131-160,173,488. For cultured cells, only the transcribed strand of CASQ1 is shown. (b) Chromatin state segmentation; Enh, enhancer chromatin; Pr, promoter chromatin; LCL, lymphoblastoid cell line; lung fib., lung fibroblasts; skeletal muscle #1 and #2, psoas muscle and an undesignated type of SkM sample, respectively; heart, left ventricle; PBMC, peripheral blood mononuclear cells. (c) Bisulfite-seq profiles for the indicated samples with blue bars above each profile indicating low-methylation regions (LMRs), which had significantly lower methylation than the rest of the genome [27]; skeletal muscle, psoas. (d) CpG islands and human/mouse conserved transcription factor binding sites. (e) DNaseI hypersensitivity peaks. (f) Human/mouse DNA sequence conservation. All tracks are from the hg19 reference genome in the UCSC Genome Browser [22] and have been aligned in this and the subsequent two figures. See text for designations of boxed regions.