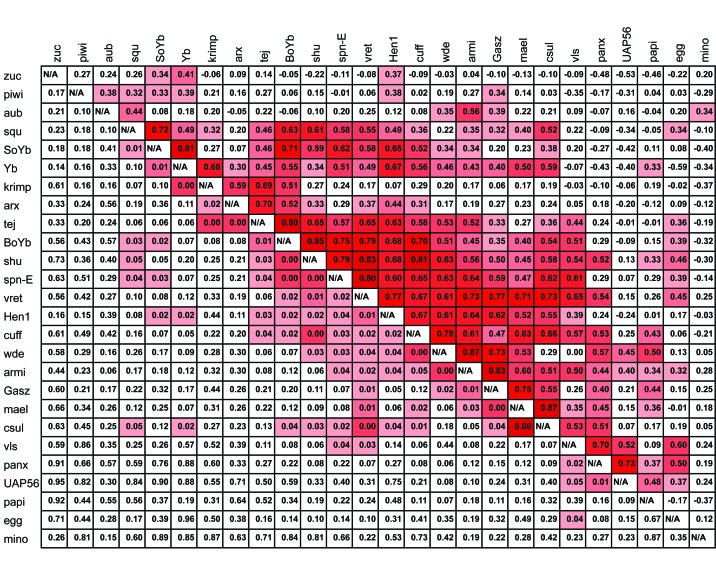
Figure 3.
Evolutionary Rate Covariation analysis of piRNA proteins across the Drosophila genus. Results were obtained from [84]. Proteins in the middle of the table share the greatest signature of co-evolution. Evolutionary Rate Covariation (ERC) analysis measures the degree to which changes in one protein (relative to background) are correlated with changes in another (also relative to background). The analysis is performed by estimating branch specific amino acid divergence across a phylogeny. Here, branch specific amino acid divergence was estimated for 12 sequenced species of the Drosophila genus. Significance is measured relative to the genomic background of all pairwise covariation estimates. Some proteins are not available due to lack of clear orthologs in divergent species. Above the diagonal: ERC values. Values range from -1 to 1, with values closer to 1 indicate higher levels of covariation. Red intensity above diagonal scales with strength of correlation. Below the diagonal: P values. P values were determined empirically relative to background. Red intensity below diagonal scales with degree of significance. Significant co-variation across the piRNA machinery is demonstrated by a Z-score of observed P values being equal to -15.4 (P << .001).