Microtubules contribute to key cellular processes and are composed of αβ-tubulin heterodimers. Neurons in Caenorhabditis elegans express cell type–specific isoforms in addition to a shared repertoire and rely on tubulins for neurite outgrowth. Isoform function varies between in vivo and in vitro contexts.
Abstract
Microtubules contribute to many cellular processes, including transport, signaling, and chromosome separation during cell division. They comprise αβ‑tubulin heterodimers arranged into linear protofilaments and assembled into tubes. Eukaryotes express multiple tubulin isoforms, and there has been a longstanding debate as to whether the isoforms are redundant or perform specialized roles as part of a tubulin code. Here we use the well‑characterized touch receptor neurons (TRNs) of Caenorhabditis elegans to investigate this question through genetic dissection of process outgrowth both in vivo and in vitro. With single‑cell RNA-seq, we compare transcription profiles for TRNs with those of two other sensory neurons and present evidence that each sensory neuron expresses a distinct palette of tubulin genes. In the TRNs, we analyze process outgrowth and show that four tubulins (tba‑1, tba‑2, tbb‑1, and tbb‑2) function partially or fully redundantly, whereas two others (mec‑7 and mec‑12) perform specialized, context‑dependent roles. Our findings support a model in which sensory neurons express overlapping subsets of tubulin genes whose functional redundancy varies among cell types and in vivo and in vitro contexts.
INTRODUCTION
Shortly before the discovery that microtubules (MTs) are composed of tubulin subunits (Mohri, 1968), investigators found differences in the chemical properties and thermal stability of various MTs (Behnke and Forer, 1967). These experiments, along with experiments showing the inducible expression of chemically distinct tubulins (Fulton and Simpson, 1976), led to the formulation of the multitubulin hypothesis, which posits that there are multiple tubulin isoforms within cells and that each one has a distinct function (Fulton and Simpson, 1976).
Early experiments in the budding yeast Saccharomyces cerevisiae argued against the multitubulin hypothesis by showing that its two α−tubulins can function redundantly (Schatz et al., 1986). Similarly, the β−tubulins of Aspergillus nidulans were also found to be interchangeable (May, 1989). In contrast, Hoyle and Raff (1990) observed isoform‑dependent function in Drosophila. In particular, they demonstrated that the function of the sole β‑tubulin isoform expressed in the male testes, β2, cannot be rescued by expression of the β3 isoform and that the β3 isoform acts in a dominant‑negative manner when coexpressed with β2 at levels exceeding 20% of the total tubulin.
The observation that most studies in isolated cells demonstrated functional redundancy, whereas the majority of studies in multicellular organisms showed isoform‑specific function, led to the question of whether isoform‑specific function may be context dependent (Ludueña, 1993; Hurd et al., 2010). In this study, we leverage the genetically tractable model organism Caenorhabditis elegans and its well-characterized and compact nervous system to address this question with respect to the role that MTs play in axon outgrowth. We use RNA-sequencing (RNA-seq) to survey the genes expressed in single identified mechanoreceptor, thermoreceptor, and chemoreceptor neurons and mine the resulting data sets to determine the tubulin genes expressed in each type of neuron. Leveraging this information, we manipulate the tubulin composition of the well‑characterized touch receptor neurons (TRNs) both in vivo and in vitro and examine the consequences of these alterations on the outgrowth of their neural processes.
C. elegans nematodes rely on six TRNs to sense gentle touch along the length of their bodies (Chalfie and Thomson, 1979; Chalfie et al., 1985). The lateral TRNs, ALML/R and PLML/R, arise embryonically, whereas the ventral neurons, AVM and PVM, arise later in larval development (Chalfie and Sulston, 1981). During development, the TRN cell bodies migrate toward their adult positions and then extend processes anteriorly, and, in the case of the PLMs, posteriorly as well (Chalfie and Sulston, 1981). The anteriorly directed PLM process grows rapidly upon hatching and then pauses in a SAX‑1/SAX‑2–dependent manner before resuming growth (Gallegos and Bargmann, 2004). This pause has been shown to be crucial for the proper tiling of the body surface (Gallegos and Bargmann, 2004).
TRN axons are packed with a distinctive bundle of 15‑protofilament microtubules that depend on the expression of MEC‑12 α‑tubulin and MEC‑7 β‑tubulin (Chalfie and Thomson, 1979, 1982; Chalfie and Au, 1989; Savage et al., 1989). These unusual microtubules are required for gentle touch sensation (Chalfie and Sulston, 1981) and to regulate gene expression (Bounoutas et al., 2011) but not for the generation of electrical currents in the TRNs in response to a mechanical stimulus (O’Hagan et al., 2005; Bounoutas et al., 2009). MEC‑12 α‑tubulin is the only C. elegans tubulin subject to acetylation at lysine 40; this modification is needed for normal touch sensation (Akella et al., 2010; Shida et al., 2010; Topalidou et al., 2012) and to constrain the number of protofilaments in each microtubule to 15 (Cueva et al., 2012).
Unlike mec‑7 and mec‑12, the expression profile of other tubulin isoforms in TRNs and their contribution to the development and function of these cells are poorly understood. Of the other eight α- and five β-tubulins in the C. elegans genome (C. elegans Sequencing Consortium, 1998; Gogonea et al., 1999), previous investigators found evidence for the expression of two other α‑tubulins in the TRNs in addition to MEC‑12: tba‑1 and tba‑2 (Fukushige et al., 1993, 1995).
Coupled with visualization of tubulin gene expression via transcriptional green fluorescent protein (GFP) fusions and clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9–mediated insertion of TagRFP-T into the genome, our study indicates that each neuron expresses a distinct palette of tubulin isoforms. The TRN mechanoreceptors express transcripts encoding three α‑ and three β‑tubulins in addition to mec‑7 and mec‑12. We analyze the effect of null mutations in a subset of these isotypes in vivo and in vitro and demonstrate that mec‑7 is required for the wild-type outgrowth of TRN processes in vivo and in vitro, whereas mec‑12 plays a partially redundant role with tba-1 in vivo and is required for wild-type outgrowth in in vitro.
RESULTS
To study the effects of tubulin isoforms on TRN process outgrowth, we first sought to understand the contribution of microtubules to the outgrowth of TRNs in vivo. Previous work established that growing C. elegans on medium containing the microtubule‑destabilizing drug colchicine was sufficient to eliminate microtubules in the lateral TRNs (Chalfie and Thomson, 1982; Bounoutas et al., 2011). We thus examined the effect of colchicine on TRNs of worms expressing one of two TRN‑specific GFP transgenes, uIs30 or uIs31 (O’Hagan et al., 2005; Bounoutas et al., 2009). As expected from prior work (Chalfie and Thomson, 1982), colchicine impaired touch sensation: untreated animals responded to an average of 8.2 ± 0.16 (mean ± SEM, n = 75 animals) of 10 trials, whereas colchicine‑treated animals responded to 2.3 ± 0.17 of 10 trials (mean ± SEM, n = 75).
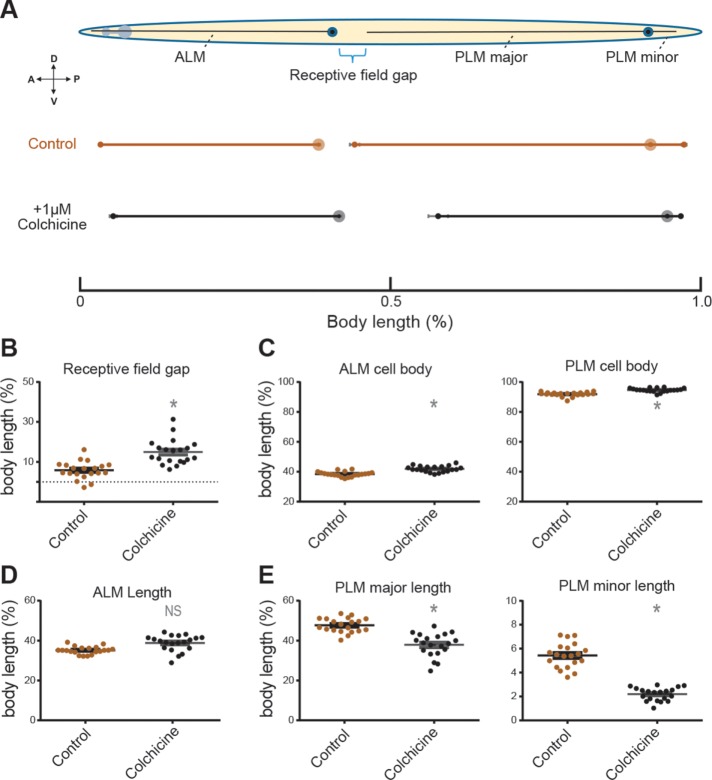
Within each cohort of animals, the position (normalized to body length) of the ALM and PLM cell bodies and the endpoints of their processes were highly stereotyped (Figure 1). The average lengths of the anterior processes of ALM and PLM were similar to those reported previously in untreated, wild-type animals (Gallegos and Bargmann, 2004; Shida et al., 2010; Petzold et al., 2013): 382 ± 12 μm (mean ± SEM, n = 20) and 498 ± 18 μm (mean ± SEM, n = 20) for ALM and PLM, respectively. Colchicine treatment shifted PLM cell bodies posteriorly and shortened both the anterior and posterior PLM processes (Figure 1, A, C, and E). The defect in PLM positioning and outgrowth resulted in an increase in the receptive field gap between the ALM and PLM neurons (Figure 1, A and B). Colchicine treatment also displaced the ALM cell body posteriorly (Figure 1C) but did not appear to affect the length of the anteriorly directed ALM process (Figure 1D). The origin of this positioning defect is unclear. ALM but not PLM cell bodies migrate a considerable distance posteriorly during development (Sulston and Horvitz, 1977; Manser and Wood, 1990), suggesting that colchicine treatment may affect posteriorly directed cell migrations. Taken together with prior work establishing that colchicine-treated animals lack 15‑protofilament microtubules in both ALM and PLM (Chalfie and Thomson, 1982), these results show that disrupting MTs has surprisingly mild effects on the extension of TRN processes in vivo.
FIGURE 1:
Pharmacological disruption of microtubules disrupts PLM length in vivo. (A) Normalized results from average of 20 control uIs31[mec‑17p::gfp] and colchicine‑treated animals grown from embryos for 3 d. Body length measured from mouth to tail. Dark circles indicate cell bodies. Anterior is left, ventral is down. (B–E) Growth and cell position parameters from all 20 animals in each treatment group expressed as a fraction of the body length. *p < 0.05 as determined by t test for a given parameter between control and colchicine‑treated groups.
TRN gene expression profiling
Having established that microtubules contribute to the outgrowth of the posterior TRNs in vivo, we sought to identify the tubulin genes that are expressed in these TRNs. To reach this goal, we used single‑cell RNA-seq to generate gene expression profiles of PLM neurons harvested from late L4 larvae. We took two steps to differentiate between tubulin genes expressed specifically in the TRNs and those expressed more broadly. First, we compared expression levels to a data set reported previously for mixed‑stage C. elegans larvae (Schwarz et al., 2012). Second, we generated gene expression profiles for two other neurons: a chemosensory neuron, ASER, and a thermosensory neuron, AFD. We detected expression of 5086 protein‑coding genes in PLM neurons (Figure 2A and Supplemental Tables S3 and S4) and similar numbers in AFD and ASER neurons (5824 and 4601 genes, respectively) isolated from animals late in larval development or in early adulthood (Figure 2 and Supplemental Tables S3 and S4). These results are similar to those reported for other neurons isolated from larvae and analyzed by RNA-seq (Spencer et al., 2014). Thus, on average, individual classes of C. elegans neurons appear to express ∼5000 genes, or one‑fourth of the genome.
FIGURE 2:
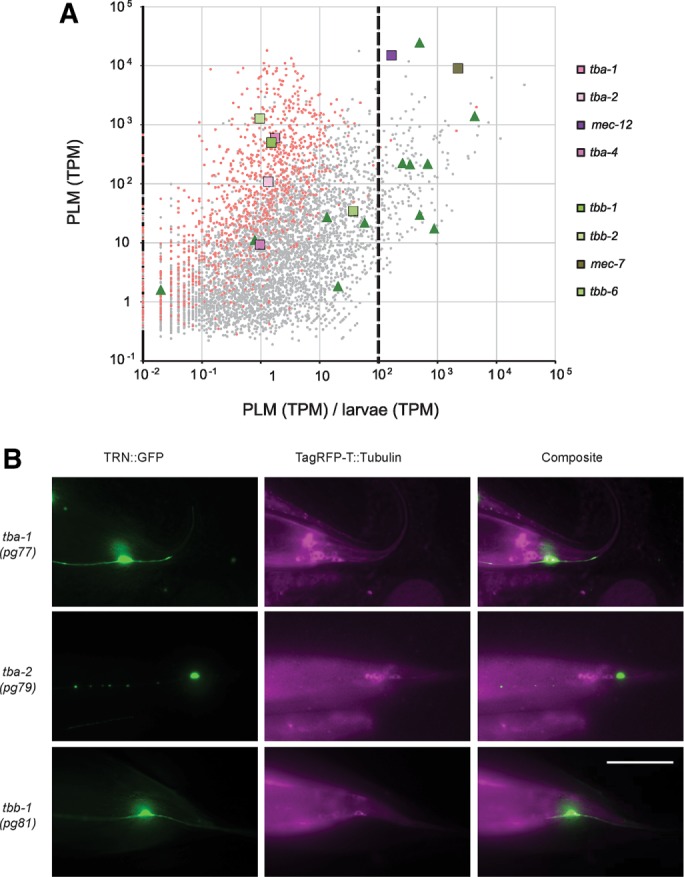
Tubulin isoform expression in PLM. (A) Expression of 5099 C. elegans genes (5086 protein coding and 13 ncRNA coding) that exhibited above‑background expression in PLM (Materials and Methods and Supplemental Table S4). Gene expression, measured in TPM, vs. expression in PLM relative to expression in whole larvae. Each gray or red dot represents a single gene expressed in PLM; red dots are 1060 housekeeping genes (Schwarz et al., 2012). Genes encoding eight tubulins are highlighted as larger squares and those encoding 12 nontubulin mec genes are shown as larger green triangles. The dashed vertical line highlights 100-fold more transcripts in PLM than in whole larvae. (B) Coexpression of α-tubulin and β-tubulin TagRFP-T protein fusions and uIs31 TRN::GFP transcriptional fusion. Left, uIs31(mec‑17p::gfp) showing PLM. Center, TagRFP‑T tubulin fusion protein. Right, composite image. TagRFP‑T was false colored as magenta for enhanced visibility. Expression of isoforms in TRNs is seen as white in the composite overlap of uIs31 marker and TagRFP‑T::Tubulin. Images are oriented so that the anterior of the animal is on the left and the ventral side is on the bottom. Scale bar, 50 μm.
The number of genes expressed in these three sensory neurons was roughly half of that detected in mixed‑stage whole C. elegans larvae, which we used as a control for general, non–PLM‑specific gene activity (9918 genes). Of the protein‑coding genes expressed in PLM, 1060 (21%) were housekeeping genes by previously defined criteria (Schwarz et al., 2012). Conversely, 955 genes (19%) could be defined as PLM enriched, 1089 (24%) as ASER enriched, and 1203 (21%) as AFD enriched by the criterion that each gene exhibited at least 10‑fold more expression in the relevant sensory neuron than in whole larvae (Supplemental Table S4). By these criteria, PLM-enriched genes included the following genes that mutate to disrupt touch sensation (in decreasing order of expression ratios): mec‑18, mec-7, mec‑10, mec‑1, mec‑4, mec‑17, mec‑2, mec‑9, mec-12, mec‑6, mec‑14, and mec‑8. AFD-enriched genes included gcy-8, gcy-23, gcy-18, hen-1, and ttx-1, and ASER-enriched genes included gcy-22, gcy-19, and flp-6. Consistent with its long cilium, transcripts encoding subunits of the BBSome, IFT-A, and IFT-B ciliary transport complexes were enriched in ASER by an average of 27-, 88-, and 100-fold, respectively. Similar enrichment was not detected in AFD (which has a very short cilium) or PLM, which lacks ciliated structures entirely. Thus application of RNA-seq technology to single identified neurons extracted from older nematodes is sufficient to detect neuron-specific genes.
C. elegans has nine α- and six β-tubulin genes in its genome (C. elegans Sequencing Consortium, 1998; Gogonea et al., 1999). We detected transcripts for eight of these 15 tubulins in PLM neurons. In decreasing order of PLM‑specific expression, they were mec‑7, mec‑12, tbb‑6, tba‑1, tbb‑1, tba‑2, tba‑4, and tbb‑2 (Figure 2A). Because we detected only low levels of expression for tba‑4 and tbb‑6 in the PLMs (nine and 34 transcripts per million [TPM], respectively), we focused on the other tubulin isoforms in this study. In the AFD neuron, we detected nine isoforms (tba‑1, tba‑2, mec‑12, tba‑4, tbb‑1, tbb‑2, mec‑7, tbb‑4, and tbb‑6; Supplemental Figure S1), and in the ASER neuron, we detected eight (tba‑1, tba‑2, mec‑12, tba‑4, tbb‑1, tbb‑2, mec‑7, and tbb‑4; Supplemental Figure S1). These results are consistent with a previous study that found tbb‑4 expression in AFD and ASE (Hurd et al., 2010); however, in contrast with their results, we did not observe expression of tba‑9 in either ciliated neuron. In AFD and ASER, we also detected significant expression of the γ‑tubulin, tbg‑1 (Supplemental Figure S1 and Supplemental Table S4). Based on this analysis, the PLM mechanoreceptors, ASER chemoreceptors, and AFD thermoreceptors all appear to coexpress transcripts encoding three α‑tubulins (tba‑1, tba‑2, mec‑12) and three β‑tubulins (tbb‑1, tbb‑2, mec‑7).
The PLM neurons expressed at least 10‑fold more transcripts encoding all tubulins than either AFD or ASER (Figure 2A, Supplemental Figure S1, and Supplemental Table S4). This difference in total tubulin transcript expression is primarily due to the large increase in mec‑7 and mec‑12 transcript numbers observed in PLM compared with AFD and ASER. We detected ∼15,000 TPM and ∼9000 TPM for mec‑12 and mec‑7, respectively, in the PLM (Supplemental Table S4). In contrast, the highest‑expressed isoform in either AFD or ASER is tbb‑2 in AFD at 889 TPM, roughly an order of magnitude lower. After mec‑7, tbb‑2 was also the most highly expressed β‑tubulin in the PLM (Figure 2A, and Supplemental Figure S1). Similarly, tba‑1 was the most highly expressed α‑tubulin in AFD and ASER, as well as in PLM, after mec‑12.
We found expression of tbb‑4 in both AFD and ASER but not in PLM (Supplemental Figure S1). tbb‑4 was previously shown to be expressed in ciliated neurons and be required for their morphology and for behaviors that depend on these neurons (Portman and Emmons, 2004; Hurd et al., 2010; Hao et al., 2011). Collectively these data suggest that each sensory neuron has a distinct tubulin expression palette and that the 10- to 100‑fold amplification in the expression of mec‑12 and mec‑7 in the TRNs is consistent with the central role that these tubulins play in TRN development and function.
Tubulin genes expressed in TRNs
Next we sought to characterize the cells and tissues expressing the tubulin isoforms detected in TRNs. Because the expression patterns of mec‑7 and mec‑12 are well established (Chalfie and Sulston, 1981; Chalfie and Thomson, 1982; Savage et al., 1989, 1994; Hamelin et al., 1992; Fukushige et al., 1999; Bounoutas et al., 2009), we did not examine them further. Instead, we focused on tba‑1, tba‑2, tbb‑1, and tbb‑2 and used CRISPR/Cas9‑mediated gene editing to insert a fluorescent protein into the native loci as well as conventional transcriptional fusions to visualize their expression.
Using a self‑excising cassette strategy (Dickinson et al., 2013), we inserted sequences encoding TagRFP‑T into the tba‑1, tba‑2, and tbb‑1 loci, creating the pg77, pg79, and pg81 insertion alleles expressing TagRFP-T fusion proteins. (Efforts to insert the same tag into the tbb‑2 locus were unsuccessful.) In lines tagging the amino terminus of TBA‑1, TBA‑2, and TBB‑1, we detected fluorescence in body wall muscle, pharynx, intestine, and neurons in the head, tail, and ventral nerve cord. Throughout the body, TagRFP‑T fluorescence was considerably higher for tba‑1 and tbb‑1 insertions than for tba‑2. These variations in apparent expression levels could reflect our finding that tba‑2 transcript counts were lower than those for tba‑1 and tbb‑1 in all three of the neurons examined with RNA-seq (Figure 2A and Supplemental Figure S4).
To determine whether these tubulins were expressed in the TRNs, we combined the pg77, pg79, and pg81 TagRFP-T insertion alleles with uIs31, which drives expression of GFP exclusively in the TRNs. We found TBA‑1 and TBB‑1 in all six TRNs but could not detect TBA‑2 in any TRNs with this method (Figure 2B). We also generated conventional transcriptional fusions and obtained lines expressing GFP downstream of the putative promoter sequences for tba‑1, tba‑2, and tbb‑2. (Efforts to visualize expression from the tbb‑1 promoter were unsuccessful.) We detected fluorescence from expression of all three promoters throughout the body (Supplemental Figure S2), as well as in the TRNs (Supplemental Figure S3), including from the tba‑2 promoter.
In summary, we tested for TRN expression using two complementary fluorescent protein–based visualization methods. For tba‑1, fluorescence was visible both from an insertion allele and from expression of a transcriptional fusion. For tba‑2, fluorescence was not visible from the insertion allele, but it was visible from expression of a transcriptional fusion. Even though only one visualization method was successful for tba-2, tbb‑1, and tbb‑2, we suggest that these results indicate that the TRNs express tba‑1, tba‑2, tbb‑1, and tbb‑2.
mec‑7 is necessary for wild-type TRN outgrowth in vivo
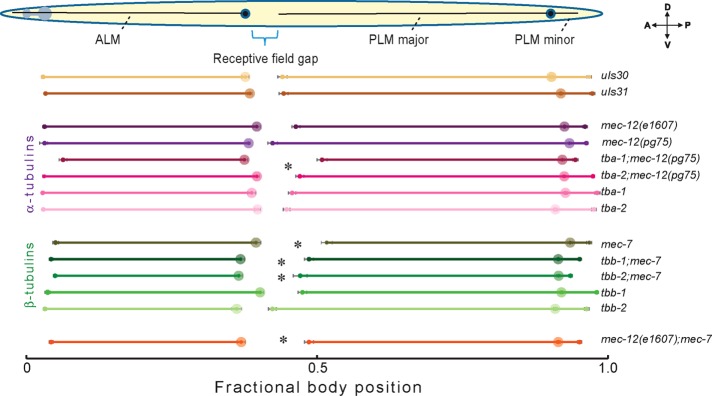
To study the contribution of different tubulin isoforms to TRN outgrowth, we created animals with putative null mutations for one or more tubulin isoforms in the presence of either uIs31 or uIs30 transgenes, which drive GFP expression with the TRN-specific promoter of the mec‑17 gene. Figure 3 compares the positions of the ALM and PLM cell bodies and their process terminations in control animals and single α‑tubulin mutants (tba‑1, tba‑2, and mec‑12), single β‑tubulin mutants (tbb‑1, tbb‑2, and mec‑7), and selected double α‑tubulin and β‑tubulin mutants. We were unable to examine tba‑1tba‑2, tba-1;tbb-1, tba-1;tbb-2, and tbb‑2tbb‑1 double mutants due to their embryonic lethality (Phillips et al., 2004; Baran et al., 2010).
FIGURE 3:
Mutation of mec‑7 or combination of mec‑12 with tba‑1 or tba‑2 mutations increases the receptive field gap. Normalized results from average of 20 animals imaged for each genotype as indicated. Anterior left, ventral down. Asterisk indicates a statistically significant increase in receptive field gap compared with respective GFP control (p < 0.05). Table 1 lists the values for all of these parameters.
Of these six tubulin genes, mec‑7 had the strongest effects on TRN process outgrowth (Figure 3 and Table 1). Loss of mec‑7 nearly tripled the length of the receptive field gap (from 4.2 ± 1% to 12.2 ± 0.8% body length), an effect that arises primarily from defects in the positioning of the PLM cell body and its process outgrowth. Animals carrying the e1607 and pg75 null alleles of mec-12 had slightly longer PLM process than wild type, but those carrying null mutations in tba‑1, tba‑2, tbb‑1, or tbb‑2 had essentially wild-type processes (Figure 3 and Table 1). Of note, tba‑1;mec‑12(pg75) double mutants had a larger receptive field gap (13.3 ± 0.9% body length) than either single mutant and comprised the only example of genetic enhancement among the double mutants we examined.
TABLE 1:
Position of the ALM and PLM cell bodies and their process lengths in vivo as a function of genotype.
| Cell body position (%) | Process length (%) | ||||||
|---|---|---|---|---|---|---|---|
| Genotype | Body length (μm) | ALM | PLM | ALM | PLM (major) | PLM (minor) | Receptive field gap (%) |
| uIs31[pmec-17::gfp] | 1148 ± 24 | 38.4 ± 0.4 | 91.9 ± 0.3 | 35.1 ± 0.4 | 47.7 ± 0.8 | 5.4 ± 0.2 | 5.8 ± 1 |
| uIs30[pmec-17::gfp] | 1127 ± 31 | 37.7 ± 0.6 | 90.3 ± 0.3 | 34.8 ± 0.5 | 46.3 ± 0.7 | 6.5 ± 0.3 | 6.4 ± 0.9 |
| uIs30; mec‑12(e1607) | 1149 ± 27 | 38.2 ± 0.5 | 93.4 ± 0.4 | 35.1 ± 0.6 | 51.1 ± 0.9 | 2.9 ± 0.2 | 4.1 ± 1 |
| mec-12(pg75)uIs31 | 1096 ± 15 | 39.6 ± 0.6 | 92.5 ± 0.5 | 36.5 ± 0.6 | 46.2 ± 0.6 | 4.0 ± 0.2 | 6.7 ± 0.9 |
| tba-1(ok1135); uIs31 | 1186 ± 20 | 38.7 ± 0.7 | 92.7 ± 0.4 | 35.9 ± 0.7 | 47.0 ± 0.6 | 5.5 ± 0.3 | 6.9 ± 0.9 |
| tba-1(ok1135); mec‑12(pg75)uIs31 | 895 ± 15 | 37.5 ± 0.6 | 92.1 ± 0.4 | 31.7 ± 1 | 41.3 ± 0.8 | 2.2 ± 0.2 | 13.3 ± 0.9 |
| tba-2(tm6948); uIs31 | 1128 ± 25 | 39.7 ± 0.5 | 91.0 ± 0.3 | 36.9 ± 0.4 | 46.2 ± 0.6 | 6.6 ± 0.4 | 5 ± 0.7 |
| tba‑2(tm6948); mec‑12(pg75)uIs31 | 1200 ± 23 | 39.6 ± 0.5 | 92.5 ± 0.3 | 36.6 ± 0.5 | 45.5 ± 0.6 | 4.9 ± 0.3 | 7.4 ± 0.7 |
| uIs31; mec-7(u142) | 1145 ± 33 | 39.5 ± 0.8 | 93.5 ± 0.4 | 34.5 ± 0.7 | 41.9 ± 0.8 | 3.3 ± 0.2 | 12.2 ± 0.8 |
| uIs30; tbb-1(gk207) | 1100 ± 21 | 40.2 ± 0.5 | 92.0 ± 0.3 | 36.7 ± 0.7 | 44.9 ± 0.7 | 6.1 ± 0.3 | 7.3 ± 0.8 |
| uIs30; tbb‑1(gk207); mec‑7(u142) | 1027 ± 32 | 36.8 ± 0.8 | 91.8 ± 0.4 | 32.1 ± 0.7 | 41.3 ± 0.9 | 3.1 ± 0.4 | 13.6 ± 1 |
| uIs30;tbb-2(gk130) | 1148 ± 20 | 36.2 ± 0.8 | 90.9 ± 0.3 | 33 ± 0.7 | 48.6 ± 0.6 | 5.4 ± 0.3 | 6.1 ± 1 |
| uIs30;tbb‑2(gk130); mec-7(u142) | 1234 ± 24 | 36.5 ± 0.6 | 91.5 ± 0.3 | 31.2 ± 0.8 | 44.5 ± 1 | 2 ± 0.2 | 10.6 ± 1.3 |
| uIs30; mec‑12(e1607); mec‑7(u142) | 1053 ± 26 | 36.9 ± 0.7 | 91.5 ± 0.3 | 32.7 ± 0.7 | 42.9 ± 0.7 | 3.7 ± 0.3 | 11.6 ± 0.8 |
| One-way ANOVA, uIs31 strains | F(6133) | 2.455 | 4.739 | 7.620 | 12.20 | 28.52 | 17.77 |
| P | 0.0277 | 0.0002 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| One-way ANOVA for uIs30 strains | F(6133) | 4.166 | 7.105 | 8.185 | 17.06 | 35.31 | 12.31 |
| P | 0.0007 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
uIs31 and uIs30 are wild-type, control animals expressing GFP exclusively in the TRNs. Data are mean ± SEM (n = 20) and reported as a percentage of body length, except for body length. One-way ANOVA was used to determine the effect of genotype and applied separately to strains containing either the uIs31 or uIs30 transgene, followed by Dunnett’s multiple comparisons test between individual genotypes and their GFP control. Values in bold are those that differed from control at the p < 0.05 level.
The minor defects in TRN process growth found in mec‑12–null mutants and tba‑1;mec‑12 double mutants are surprising for at least two reasons. First, 95% of all α‑tubulin transcripts detected in PLM encode mec‑12, and 99% of these transcripts encode either mec‑12 or tba‑1 (Supplemental Table S4). Second, mec‑12 loss-of-function mutants are expected to lack the distinctive bundle of 15‑protofilament microtubules present in wild‑type TRNs (Chalfie and Au, 1989; Cueva et al., 2012). Collectively these observations suggest that other tubulins may compensate for the loss of mec‑12 and tba‑1 and that 15‑protofilament microtubules are not essential for TRN outgrowth in vivo.
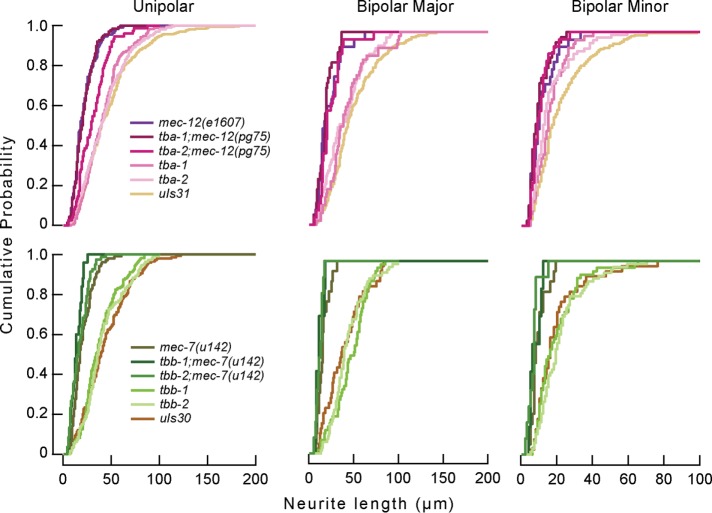
We used a 10-trial, classical gentle touch assay (Chalfie et al., 2014) to assess TRN function in single and double tubulin mutants. Consistent with earlier work (Chalfie and Sulston, 1981), we found that null mutations in mec‑7 and mec‑12, alone or in combination, caused severe defects in touch sensation (Figure 4). In contrast, touch sensation was indistinguishable from wild type in null mutants of the four other tubulins and indistinguishable from the defects seen for mec‑12 or mec‑7 in double mutants. These results are consistent with an important role for mec‑12 and mec‑7 tubulins in TRN function.
FIGURE 4:
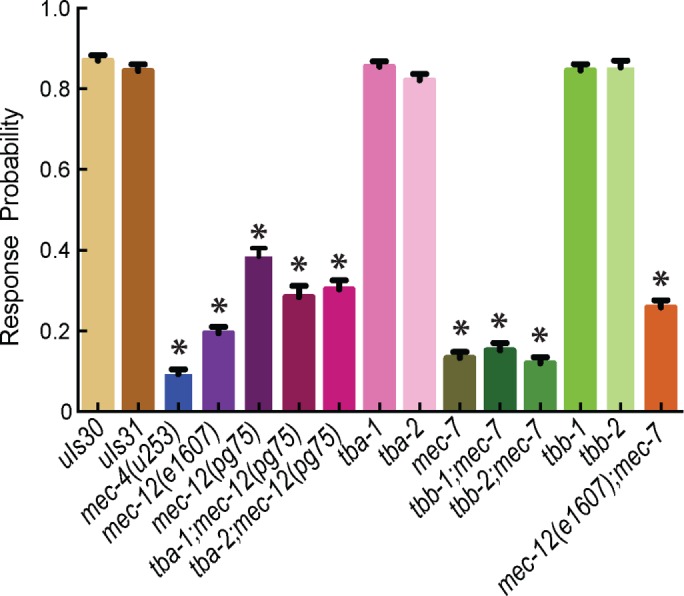
The tba‑1, tba‑2, tbb‑1, or tbb‑2 loss-of-function mutants have wild-type touch sensitivity. Results from gentle touch assays on 75 animals per genotype conducted blind to genotype on three separate days. Experimenter blinded to genotype. Statistical significance determined by omnibus ANOVA with respective GFP control, followed by Dunnett’s multiple comparisons test between individual genotypes and their GFP control. *p < 0.05.
Wild‑type TRNs extend unipolar and bipolar processes in vitro
We sought to address the potential contribution of the extracellular matrix (ECM) to process outgrowth and explore the interplay between tubulin isoforms, the ECM, and process outgrowth by isolating the TRNs from their native context. To reach this goal, we first dissociated uIs31 and uIs30 control embryos in which GFP is expressed only in the TRNs (Materials and Methods) and characterized the processes they extended in vitro. TRN morphologies observed in vitro were broadly similar to those observed in vivo (Figure 5A) and to what was reported previously (Zhang et al., 2002; Strange et al., 2007). The GFP‑labeled cells fell into two categories—unipolar and bipolar—and extended three types of processes—unipolar, major bipolar, and minor bipolar (Figure 5A). We plotted the pooled data as cumulative probability distributions when making comparisons between genotypes, as seen in Figure 6B. The geometric means for control TRNs were 38.7 μm for the unipolar processes, 41.1 μm for the bipolar major processes, and 17.5 μm for the bipolar minor processes. We observed no effect of plating density on process length in the range of (0.5–1.5) × 105 cells/dish (unpublished data). On average, all three types of processes were ∼10-fold shorter in vitro than in vivo (Figure 3 and Table 1; also see later discussion of Figure 7).
FIGURE 5:
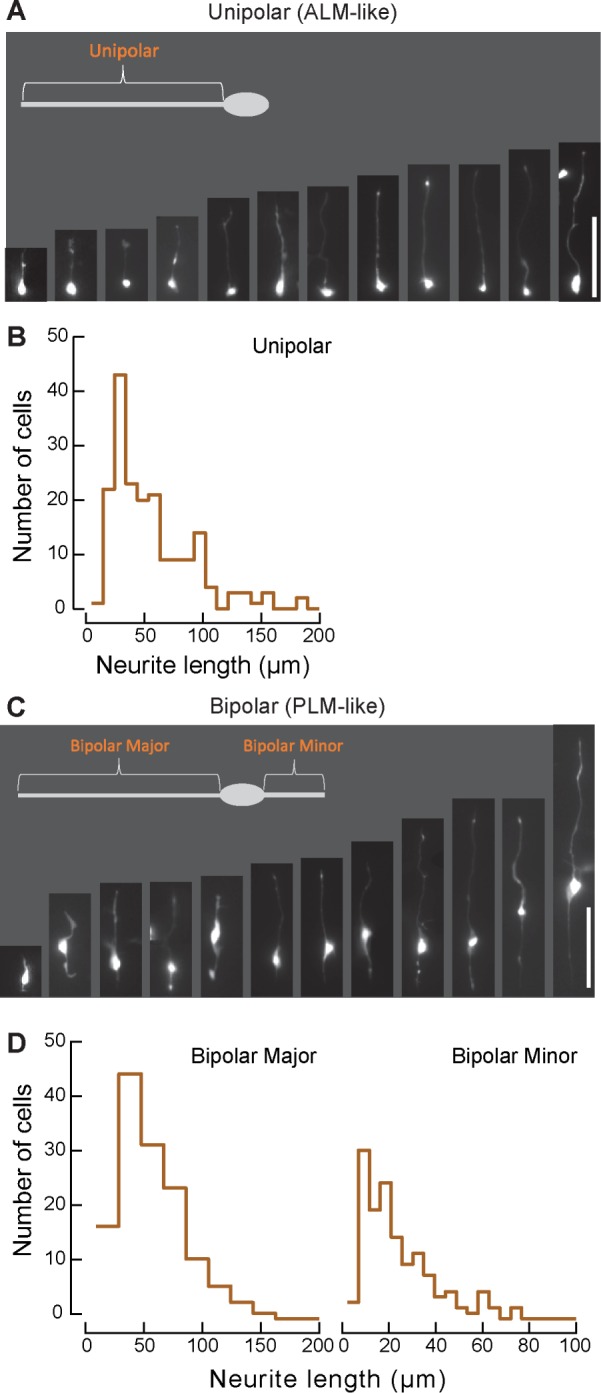
TRNs adopt similar morphologies in vitro as they do in vivo. (A, C) Example images of unipolar and bipolar cells in culture. Processes are described as unipolar for the single process on unipolar cells, as bipolar major for the large process on bipolar cells, and as bipolar minor for the small process. Scale bars, 50 μm. (B, D) Histograms of process lengths for (B) unipolar and (D) major bipolar and minor bipolar cells. Bin sizes were determined as in Shimazaki and Shinomoto (2007).
FIGURE 6:
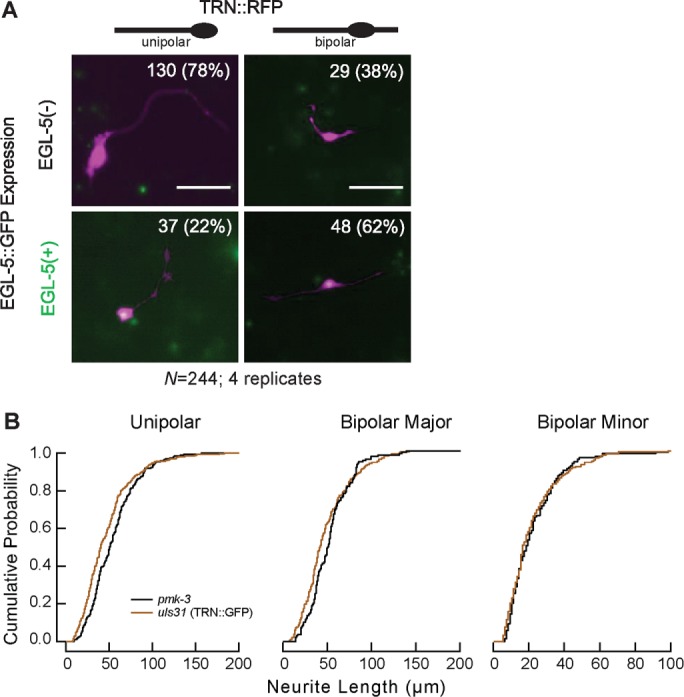
TRNs primarily adopt ALM‑like cell fate and extend processes by a pmk‑3‑independent mechanism in vitro. (A) Representative images of TRNs cultured from animals expressing TRN::RFP and EGL‑5::GFP. RFP is false‑colored as magenta and GFP as green. Left, unipolar cells; right, bipolar cells. Top, tEGL‑5(‑) cells; bottom, EGL‑5(+) cells. Morphology and EGL‑5 expression were scored manually. Scale bars, 50 μm. (B) Cumulative probability distributions for TRNs isolated from control uIs31[pmec‑17::gfp] and pmk‑3(ok169) animals. Statistical significances were determined by Kolmogorov–Smirnov tests.
FIGURE 7:
mec‑7 and mec‑12 are necessary for wild‑type process outgrowth in vitro. Cumulative probability distributions for TRNs isolated from animals of given genotypes for α−tubulin mutants (top) and β−tubulin mutants (bottom). Plots show data for the indicated mutants together with their respective controls, except for mec‑12 and mec‑7, which are plotted by tubulin family (α or β) for comparisons within class. No differences were detected between uIs30 and uIs31 controls.
Whereas in vivo process lengths have a small variance between individual animals and are normally distributed (Figure 1, B–E), processes grown in vitro exhibit substantially larger variance in their length, and their distribution is log‑normal (Figure 5B). Indeed, control neuron lengths had coefficients of variation (CVs) that were 6 and 80% for the anterior process of ALM in vivo and unipolar processes in vitro, respectively. Similarly, CV values were 8% for the anterior process of PLM in vivo and 90% for the major process of bipolar, PLM-like processes in vitro. This finding suggests that process length is tightly controlled in vivo but not in vitro.
Although TRN shapes were similar in vitro to those found in vivo, where the ALM neurons are unipolar and the PLM neurons are bipolar, we sought to determine how in vitro morphology was connected to the cell fates of ALM and PLM. To address this question, we obtained uIs115;uIs116 transgenic animals expressing red fluorescent protein (RFP) selectively in the TRNs and a GFP‑tagged EGL‑5, which is expressed in PLM but not ALM (Ferreira et al., 1999; Zheng et al., 2015a), and dissociated cells from these embryos. Based on the expression of EGL‑5::GFP, the majority of TRNs in vitro appear to adopt an ALM‑like cell fate in terms of both their unipolar morphology and lack of detectable EGL‑5 expression. Sixty‑eight percent of TRNs were unipolar; most of these neurons (78%) also did not express EGL‑5, consistent with an ALM cell fate (Figure 6A). The remaining cells were bipolar; slightly more than half (62%) expressed EGL‑5, as expected for a PLM cell fate. Given that embryos contain equal numbers of ALM and PLM cells, we performed a chi‑squared goodness-of-fit test to compare our ratios of unipolar:bipolar and EGL‑5+:EGL‑5‑ to a hypothetical 1:1 ratio. We found that the differences were significantly biased toward ALM morphology and away from EGL‑5 expression (p < 0.001). Thus embryonic TRNs grown in culture are more likely to adopt an ALM‑like cell fate, consistent with the idea that ALM is the default cell fate for TRNs (Zheng et al., 2015a).
TRN process outgrowth is pmk‑3 p38 mitogen-activated protein kinase independent
TRNs generate processes embryonically (Gallegos and Bargmann, 2004; Hilliard and Bargmann, 2006). Similar to isolation procedures for dissociated spinal ganglia (Daniels, 1972), the embryonic dissociation procedure removes neural processes such that only cell bodies are present in cultures immediately after plating. Because C. elegans neurons can regenerate after laser‑induced axotomy in vivo (Yanik et al., 2005), we reasoned that process outgrowth could recapitulate either a regenerative process or a developmental outgrowth process. As a first step toward distinguishing between these possibilities, we cultured TRNs from animals with a mutation in the p38 mitogen-activated protein kinase, pmk‑3, which is required for process regeneration (Hammarlund et al., 2009; Yan et al., 2009) but not for TRN outgrowth in vivo (unpublished data). The pmk‑3 TRNs extended processes normally in vitro; in fact, their processes were slightly longer than those of wild-type cells in the case of the unipolar cells (Figure 6B). Thus the in vitro growth process is independent of pmk‑3 and likely to differ from process regeneration in vivo. These results suggest that TRN process growth in vitro is more closely related to developmental outgrowth than it is to regeneration.
TRNs require mec‑7 and mec‑12 for outgrowth in vitro
Next we tested whether microtubules were required for outgrowth in vitro by treating control cultures with 1 μM colchicine. Although GFP-tagged cell bodies were readily detected in colchicine-treated cultures, we did not detect processes in three independent replicates. In contrast, cultures treated side by side with vehicle (dimethyl sulfoxide [DMSO]) had processes that were qualitatively and quantitatively similar to untreated controls—the geometric means of their lengths were 41.5 μm for unipolar, 42.3 μm for major bipolar, and 16.3 μm for minor bipolar. Thus colchicine treatment has stronger effects on TRN process formation in vitro than it does in vivo (Figure 1).
Next we prepared cultures from the tubulin mutant strains that we examined in our in vivo experiments. Mutations in tbb‑1 and tbb‑2 did not affect the distribution of process lengths. Similar to our observations in vivo, however, mec‑7 caused a substantial shift in the distributions of process lengths to shorter distributions. Whereas only PLM was affected in vivo, both unipolar and bipolar cells were affected in vitro. Process lengths in tbb‑1;mec‑7 and tbb‑2;mec‑7 double mutants were similar those found in mec‑7 single mutants. In contrast with our in vivo observations (Figure 3), we found that loss of mec‑12 caused a strong reduction in the lengths of both unipolar and bipolar cells (Figure 7). Process lengths in tba‑1;mec‑12 and tba‑2;mec‑12 double mutants were not significantly different from those in mec‑12 single mutants, however. The differential effect of loss of mec‑12 function in vivo and in vitro indicates that the roles of the tubulin isoforms crucially depend on their cellular context.
DISCUSSION
Animal genomes harbor multiple genes encoding α‑ and β‑tubulins that coassemble to form microtubules essential for the growth of long, thin axons and dendrites in the nervous system. Although highly conserved overall, variable portions of α‑ and β‑tubulins diverge within genomes, and this finding fostered the multitubulin hypothesis. Our results support a model in which each neuronal type expresses a distinctive palette of tubulins, and some tubulin isoforms have evolved for significant redundancy, whereas others have acquired specialized roles essential for the function of the particular neuron type. Whereas redundancy among tubulin isoforms defends neurons against mutation of single tubulin genes, specialization enables the differentiation of microtubule structures between neuronal subtypes.
For example, the α-tubulins TBA-1 and TBA-2 are individually dispensable for the outgrowth and function of the touch receptor neurons, whereas MEC-12 is important for sensory function of TRNs and probably plays a role in neurite outgrowth that is partially compensated by TBA-1 in vivo but revealed in vitro. Similarly, the β-tubulins TBB-1 and TBB-2 are individually dispensable for process outgrowth, but loss of MEC-7 causes sensory defects and impairs outgrowth in living animals and shortens neurites in culture. The TRNs are not the only cells that rely on the TBA-1 and TBA‑2 α-tubulins and the TBB-1 and TBB-2 β-tubulins. These four tubulins cooperate in a partially redundant manner to enable mitosis and embryogenesis (Phillips et al., 2004). The TBA-1 α-tubulin, along with the TBB‑1 and TBB-2 β-tubulins, also contribute to motor neuron axon outgrowth and guidance (Baran et al., 2010). We infer from these results that TBA-1 and TBA-2 α-tubulins and TBB-1 and TBB-2 β-tubulins are incorporated not only into microtubules forming mitotic spindles, but also into microtubules that regulate process outgrowth in neurons.
Because the lateral TRNs assume stereotyped shapes and sizes in vivo and arise in the embryo, they provide a fruitful basis for understanding which features of neurite outgrowth are preserved when neurons are dissociated from embryos and grown in vitro. For instance, most cultured TRNs have simple bipolar or monopolar shapes similar to those found in vivo. Neurite lengths are far more variable in vitro than they are in vivo, however, as evidenced by an ∼10-fold higher coefficient of variation in vitro. More than half of the lateral TRNs adopt an ALM-like, EGL-5-negative cell fate in vitro. In addition, loss of MEC-12 α-tubulin shortens neurites in vitro but not in vivo. One important factor that could be linked to all of these features is the presence of native ECM in vivo and its absence in vitro. For instance, the ECM might compensate for defects in microtubules, as seen in colchicine-treated model neurons cocultured with an ECM-like material (Lamoureux et al., 1990). Whereas cell–cell and cell–matrix signaling is dispensable for outgrowth under many conditions, it could also promote growth, provide a reliable stop-growth signal, and promote expression of egl-5.
Tubulins that function redundantly in neurons could enable the divergence of tubulin genes and give rise to special-purpose tubulins serving neuron-specific functions. In humans, mutations in tubulin genes have been linked to severe defects in brain development or tubulinopathies (Romaniello et al., 2015; Chakraborti et al., 2016). Although most tubulinopathies result from missense mutations (Chakraborti et al., 2016), loss of special-purpose tubulins could alter axon guidance and outgrowth, as found for the loss of MEC‑7 in C. elegans TRNs. Our results suggest that understanding the interplay between cellular context and tubulin isoform function may offer new insights into how neurons regulate a wide array of growth phenotypes from a pool of similar building blocks.
MATERIALS AND METHODS
C. elegans strains and genetics
We used standard techniques (Brenner, 1974) to maintain strains, which were obtained from the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN), the Japanese Knockout Consortium (FXO6948), as a generous gift from C. Zheng and M. Chalfie (strain TU4008; Department of Biological Sciences, Columbia University, New York, NY), or created for the purposes of this study. Supplemental Table S1 lists the strains used in this study and their provenance. To visualize TRNs in vivo and in vitro, we used transgenes uIs31[mec‑17p::gfp]III (O’Hagan et al., 2005) or uIs30[mec‑17p::gfp]I (Bounoutas et al., 2009), which express GFP selectively in the TRNs. To identify egl‑5‑expressing TRNs in dissociated cell cultures, we used the uIs115[mec‑17p::rfp] and uIs116[egl‑5p::egl‑5::gfp] transgenes, which selectively label TRNs and EGL-5 expressing cells, respectively (Zheng et al., 2015b).
CRISPR/Cas9‑mediated gene disruption
We used CRISPR/Cas9‑mediated mutagenesis to create double tubulin knockouts and combine tubulin gene loss of function with transgenes labeling the TRNs with GFP. In particular, we generated pg75, a new mec‑12 loss‑of‑function allele, in the uIs31 background as follows. (Both mec‑12 and the uIs31 TRN::GFP transgene are located on chromosome III.) We injected purified in vitro–generated CRISPR/Cas9 complexes with sgRNA against the target sequence (5′‑GCAGTTTGTCTGCTTTTCCG‑3′) into the gonads of adult TU2769 uIs31[mec‑17p::gfp] hermaphrodites, essentially as described by Paix et al. (2015). The sgRNA was generated by in vitro transcription of a short PCR product containing the target sequence. Recombinant Cas9 was purchased from PNA Bio (CP01; Newbury Park, CA). Individual mechanosensory abnormal (Mec) F2 progeny were isolated, and their offspring were genotyped to characterize the molecular defect present in the mec‑12 gene. The pg75 allele corresponds to an eight‑nucleotide (nt) deletion of TCCGTGGA near the 5′ end of the target site, resulting in a stop codon at amino acid position 317. The pg75 allele failed to complement the previously identified null allele mec‑12(e1607) (Fukushige et al., 1999) and was recessive to the wild‑type allele in N2. pg75/e1607 transheterozygotes responded to 1 of 10 touches (n = 5), and pg75/+ heterozygotes responded to 8.5 of 10 touches (n = 5). The latter is comparable to the results for the uIs30 and uIs31 markers alone (Figure 4). The pg75 allele was outcrossed twice to minimize the contribution of off‑target mutations before using the strain to generate tba‑1(ok1135)I; mec‑12(pg75)uIs31[mec‑17p::gfp]III and tba‑2(tm6948)I;mec‑12(pg75)uIs31[mec‑17p::gfp]III double mutants.
CRISPR/Cas9‑mediated promoter and protein fusions
To visualize the cells and tissues expressing tba‑1, tba‑2, and tbb‑1, we adopted the CRISPR/Cas9‑mediated genome insertion strategy reported by Dickinson et al. (2013) and obtained GN675 pg77[TagRFP‑T::tba‑1]; uIs31, GN683 pg79[TagRFP‑T::tbb‑1]; uIs31, and GN688 pg81[TagRFP‑T::tba‑2]; uIs31 (Supplemental Table S1). (Attempts to insert sequences encoding TagRFP‑T into the tbb‑2 locus using this strategy were not successful.) For each gene, we generated an sgRNA construct based on the pDD162 plasmid (47549; Addgene, Cambridge, MA) and a homology repair construct that was based on the pDD284 plasmid (66825; Addgene). Briefly, a homology repair construct was generated by PCR with ∼500 base pairs of homologous sequences both 5′ and 3′ of the target insertion site flanking the self‑excising cassette; this PCR product was then inserted into the pDD284 plasmid using the NEB Hi‑Fi Assembly kit (NEB, Ipswich, MA). We created a Cas9‑sgRNA construct by inserting a 20–base pair target sequence into the pDD162 plasmid using the Q5 Site‑Directed Mutagenesis kit (NEB). These two constructs were injected into the gonads of young adult hermaphrodites at 10 ng/μl (homology repair) and 50 ng/μl (Cas9 construct). We also injected 50 ng/μl mg166 (unc‑122p::RFP), 5 ng/μl pCFJ104 (myo‑3p::mCherry::unc-54utr), and 2.5 ng/μl pCFJ90 (myo‑2p::mCherry::unc-54utr) as coinjection markers. Putative transgenic animals were cultivated at 25°C for 3 d; hygromycin was then added to the plates to a final concentration of 250 μg/ml to select for transgenic animals and putative knock‑in lines. After 3–5 d of hygromycin selection, we screened for animals that exhibited a strong Rol phenotype, hygromycin resistance, and a lack of coinjection markers as markers for successful insertion into the genome near the PAM site identified for each gene. To generate animals expressing protein fusion constructs, five L1s with the promoter fusion insertions obtained as described were picked to each of three nematode growth medium plates and subjected to a 34°C heat shock for 4 h and then placed back at 25°C. After 3 d, we screened for animals that were non‑Rol to obtain the fluorescent protein::tubulin fusion animals.
Tubulin promoter cloning methods
The tba-1 promoter sequence used in plasmid pRO133 was 1144 base pairs upstream of the genomic tba-1 start codon. The tba-2 promoter sequence used in plasmid pRO134 was 1018 base pairs upstream of the genomic tba-2 start codon. The tbb-2 promoter sequence used in plasmid pRO137 was 867 base pairs upstream of the genomic tbb-2 start codon. pRO133, pRO134, and pRO137 were made by fusing promoter sequences to the gfp coding sequence and unc-54 3′ untranslated region using Gibson Assembly (Gibson et al., 2009).
All tubulin promoter transgenes were made by germline injection transformation of strain PS2172 [pha-1(e2123) III; him-5(e1490)V] with an injection mix containing the following DNA: tubulin promoter gfp plasmid at 20 ng/μl, and pha-1(+) rescue plasmid (pBX; Miyabayashi et al., 1999) at 50 ng/μl and unc-122p::rfp plasmid at 50 ng/μl as transformation markers. These transgenics were selected by maintaining animals at ≥20°C, the restrictive temperature for the pha-1(e2123) mutation.
Single‑neuron RNA-seq
Microdissection and single‑cell reverse transcription (RT)–PCR of individual PLM, AFD, and ASER neurons were performed essentially as described (Schwarz et al., 2012). In particular, we dissected and harvested individual GFP+ neurons from either L4 or adult animals, separately amplified their transcripts with RT‑PCR, and purified and quantitated the RT‑PCR products. To determine consensus expression patterns for each neuron type, we pooled aliquots (of varying volume but constant DNA content) from purified and quantitated individual RT‑PCR products before performing RNA-seq on the pools. For PLM, we pooled three aliquots; for AFD, seven aliquots; and for ASER, seven aliquots. For all three neuronal types, single‑end 50‑nt RNA-seq was performed on an Illumina HiSeq 2000 (San Diego, CA). To identify so‑called housekeeping genes and genes primarily active outside the nervous system, we compared results from PLM, AFD, and ASER to published single‑end 38‑nt RNA-seq data from mixed‑stage whole C. elegans hermaphrodite larvae (Schwarz et al., 2012).
Reads were quality filtered as follows: neuronal reads that failed Chastity filtering were discarded (Chastity filtering had not been available for the larval reads); raw 38‑nt larval reads were trimmed 1 nt to 37 nt; all reads were trimmed to remove any indeterminate (“N”) residues or residues with a quality score of <3; and larval reads that had been trimmed <37 nt were deleted, as were neuronal reads that had been trimmed <50 nt. This left 19,316,855–27,057,771 filtered reads for analysis of each neuronal type versus 23,369,056 filtered reads for whole larvae (Supplemental Table S2).
We used RSEM version 1.2.17 (Li and Dewey, 2011) with bowtie2 version 2.2.3 (Langmead and Salzberg, 2012) and SAMTools version 1.0 (Li et al., 2009) to map filtered reads to a C. elegans gene index and generate read counts and gene expression levels in TPM. To create the C. elegans gene index, we ran RSEM’s rsem‑prepare‑reference with the arguments “–bowtie2 –transcript‑to‑gene‑map” on a collection of coding DNA sequences (CDSes) from both protein‑coding and non–protein‑coding C. elegans genes in WormBase release WS245 (Harris et al., 2014). We obtained the CDSes from ftp://ftp.sanger.ac.uk/pub2/wormbase/releases/WS245/species/c_elegans/PRJNA13758/c_elegans.PRJNA13758.WS245.mRNA_transcripts.fa.gz and ftp://ftp.sanger.ac.uk/pub2/wormbase/releases/WS245/species/c_elegans/PRJNA13758/c_elegans.PRJNA13758.WS245.ncRNA_transcripts.fa.gz.
For each RNA-seq data set of interest, we computed mapped reads and expression levels per gene by running RSEM’s rsem‑calculate‑expression with the arguments “–bowtie2 ‑p 8 –no‑bam‑output –calc‑pme –calc‑ci –ci‑credibility‑level 0.99 –fragment‑length‑mean 200 –fragment‑length‑sd 20 –estimate‑rspd –ci‑memory 30000.” These arguments, in particular “–estimate‑rspd,” were aimed at dealing with single‑end data from 3′‑biased RT‑PCRs; the arguments “–phred33‑quals” and “–phred64‑quals” were also used for the neuronal and larval reads, respectively. We computed posterior mean estimates (PMEs) both for read counts and for gene expression levels and rounded PMEs of read counts down to the nearest lesser integer. We also computed 99% credibility intervals (CIs) for expression data so that we could use the minimum value in the 99% CI for TPM as a reliable minimum estimate of a gene’s expression (minTPM).
We observed the following overall alignment rates of the reads to the WS245 C. elegans gene index: 50.71% for the PML read set, 28.42% for the AFD read set, 31.13% for the ASER read set, and 76.41% for the larval read set (Supplemental Table S2). A similar discrepancy between lower alignment rates for hand‑dissected linker-cell RNA-seq reads versus higher alignment rates for whole-larval RNA-seq reads was previously observed and found to be due to a much higher rate of human contaminant RNA sequences in the hand‑dissected linker cells (Schwarz et al., 2012). We defined nonzero, above‑background expression for a gene in a given RNA-seq data set by that gene having a minimum estimated expression level (termed minTPM) of at least 0.1 TPM in a credibility interval of 99% (i.e., ≥ 0.1 minTPM). The numbers of genes being scored as expressed in a given neuronal type (PLM, AFD, or ASER) above background levels are given in Supplemental Table S3 for various data sets. Other results from RSEM analysis are given in Supplemental Table S4.
We annotated C. elegans genes and the encoded gene products in several ways (Supplemental Table S4). For the products of protein‑coding genes, we predicted classical signal and transmembrane sequences with Phobius 1.01 (Käll et al., 2004), regions of low sequence complexity with pseg (SEG for proteins) from ftp://ftp.ncbi.nlm.nih.gov/pub/seg/pseg (Wootton, 1994), and coiled‑coil domains with ncoils from www.russelllab.org/coils/coils.tar.gz (Lupas, 1996). PFAM 27.0 protein domains from PFAM‑A (Finn et al., 2014) were detected with HMMER 3.0/hmmsearch (Eddy, 2009) at a threshold of E ≤ 10‑5. The memberships of genes in orthology groups from eggNOG 3.0 (Powell et al., 2012) were extracted from WormBase WS245 with the TableMaker function of ACEDB 4.3.39. Genes with likely housekeeping status were as identified described (Schwarz et al., 2012).
Embryonic cell culture
We isolated cells from C. elegans embryos and maintained them in mixed cultures as described (Strange et al., 2007). Briefly, we treated 15 6‑cm plates full of synchronized adult animals with sodium hypochlorite (Sulston and Hodgkin, 1988) to obtain an ∼50-μl pellet of embryos. These embryos were washed with egg buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.3) and separated from debris using sucrose gradient centrifugation. The embryos were then digested with chitinase for 85 min (1 U; C7809; Sigma-Aldrich, St. Louis, MO), resuspended in egg buffer (1 ml), triturated (15 times) with a micropipette with the tip pressed against the inside of a microcentrifuge tube, and filtered through a 5‑μm syringe filter. The resulting cells were counted with a hemocytometer, resuspended in nematode culture medium (L‑15 without phenol red, 10% heat‑inactivated fetal bovine serum, 50 U/ml penicillin, and 50 mg/ml streptomycin), and plated on glass‑bottomed Petri dishes (MatTek, Ashland, MA). Culture dishes were pretreated with peanut lectin (0.5 mg/ml) solution for 17 min. Cells density was adjusted to 500,000–1,000,000 cells/ml, and 50,000–100,000 cells were plated on each dish. Cells were maintained in culture at room temperature for 3 d before analysis.
Imaging TRNs
All of the images, aside from the whole-worm tubulin expression images (Supplemental Figure S2), were captured on an inverted Nikon Eclipse Ti‑E microscope (Nikon, Melville, NY) equipped with a PlanApo 100×/1.4 numerical aperture (NA) oil (egl‑5 imaging), a PlanApo 60×/1.4 NA oil (CRISPR tubulin expression imaging), a PlanApo 40×/0.95 NA air (in vitro length measurements), a PlanFluor 40×/1.3 NA oil (in vitro length measurements), or a PlanApo 20×/0.75 NA air (in vivo length measurements) objective (all objectives manufactured by Nikon). For most in vivo imaging, animals were immobilized on 5% agarose pads as reported by Kim et al. (2013), with the exception of the whole-worm tubulin expression imaging, for which the animals were anesthetized with 10 mM levamisole and mounted on agar pads for imaging at room temperature. Images were captured with a scientific complementary metal-oxide semiconductor camera (Neo 5.5; Andor Technology, Belfast, Northern Ireland) controlled by the Micromanager software package (Edelstein et al., 2014) with 4 by 4–pixel binning or, in the case of the whole-worm expression imaging, a Zeiss Axio Imager.D1M (Zeiss, Oberkochen, Germany) with a Retiga-SRV Fast 1394 digital camera (Q-Imaging, Surrey, Canada) or a Zeiss Axioplan2 microscope with a Cascade 512B (Photometrics, Tuscon, AZ) charge-coupled device camera, both of which are equipped with 10, 63 (NA 1.4), and 100× (NA 1.4) oil-immersion objectives.
GFP- or RFP-expressing TRNs were classified as either unipolar or bipolar before analysis. Cells with either one process or with two processes on which the second process was less than one cell body length were classified as unipolar. Cells with two processes that were both greater than one cell body length were classified as bipolar. Cultures also included TRNs with irregular spider web morphologies, but these were rare (<10%) and not analyzed in detail. Process lengths were measured with the segmented line tool in Fiji (Schindelin et al., 2012) after a calibration using a micrograph of a stage micrometer. The scaling conversions were (in pixels/μm) 20× air (0.78), 40× air (1.52), 40× oil (1.54), 60× oil (2.34), and 100× oil (3.80). Animals were measured from the opening of the mouth to the point at which the tail narrowed to a thin, hair‑like structure, due to the difficulty of accurately determining the final termination point of the tail. Thus our total worm length measurements are systematically smaller than the actual length of the animal from the mouth to the tip of the tail. For cell‑fate characterization, we performed in vitro cell isolations on TU4008 as described. TRNs were classified according to their morphology (unipolar or bipolar) and the presence or absence of EGL‑5::GFP.
Drug treatments
Animals were exposed to colchicine throughout development by placing eggs on standard growth plates supplemented with colchicine (1 mM) as described (Chalfie and Thomson, 1982). This concentration was below the 2 mM threshold at which animals become uncoordinated or lethargic (Chalfie and Thomson, 1982). Cultured cells were exposed to colchicine (1 μM) via the culture medium. Colchicine was prepared as stock solution in DMSO (40 mM), and its effects were compared with vehicle (DMSO) alone. Colchicine and DMSO were obtained from Sigma-Aldrich.
Touch assays
Touch sensitivity was assayed as described (Chalfie et al., 2014). Briefly, adult hermaphrodites (aged 1 d after their final larval molt) were brushed with an eyelash hair glued to a thin wooden rod. Each animal was tested with 10 stimuli: five to the anterior of the animal and five to the posterior. Responses were considered positive if they elicited either a pause or a reversal. For each genotype tested, 75 bleach‑synchronized animals were assayed in cohort of 25 animals across three replicates. All assays were conducted blinded to genotype or drug treatment.
Statistical methods
Statistical analyses were performed with PRISM (GraphPad Software, La Jolla, CA). Process length distributions, both in vivo and in vitro, were tested for normality using the Shapiro–Wilks test. For in vivo analysis of process length, we used the following statistical tests: for colchicine treatment experiments, cell parameters were compared with controls using a t test. In the case of the in vivo TRN imaging experiments or the gentle touch assays, a one‑way analysis of variance (ANOVA) was used to compare the mutant groups to their GFP marker controls. In instances in which the results of the ANOVA rejected the null hypothesis, a post hoc Dunnett’s test was used to compare each mutant or treatment group against the GFP marker control.
For in vitro analysis of neuron morphology and egl‑5 expression, we used the following statistical tests: comparisons of in vitro cell morphology and egl‑5 expression were tested using a chi‑squared goodness‑of‑fit test versus an expected ratio of 50:50 unipolar:bipolar or 50:50 GFP(+):GFP(–), respectively. Distributions of in vitro process lengths were compared using the Kolmogorov–Smirnov test to their GFP controls. Bin sizes for histograms of in vitro data were calculated as in Shimazaki and Shinomoto (2007) by minimizing the total error between the histogram and the estimated underlying distribution of the data.
Data availability
Sequence files for raw RNA-seq reads have been deposited in the National Center for Biotechnology Information Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) under the accession numbers SRR3481679 (PLM neuron pool), SRR3481680 (AFD neuron pool), and SRR3481678 (ASER neuron pool).
Supplementary Material
Acknowledgments
We thank Beth Pruitt and Maria Gallegos for discussion and feedback, Zhiwen Liao for numerous microinjections, Chloé Girard for technical consultation, the Japanese Knockout Consortium, Martin Chalfie, and Chaogu Zheng for strains, Igor Antoshechkin at the Jacobs Genomics Laboratory (California Institute of Technology, Pasadena, CA) for RNA-seq, WormBase, and the Goodman, Pruitt, and Dunn groups for insightful discussions regarding this work. This work was funded by National Institutes of Health Research Grants K99NS089942 (to M.K.), R01NS047715 (to M.B.G.), R01GM084389 (to P.W.S.), and R01DK059418 and R56DK074746 (to M.M.B.), New Jersey Commission on Spinal Cord Research Grant CSCR15IRG014 (to R.O.), and a Stanford Graduate Fellowship and National Institutes of Health Cellular and Molecular Biology Training Program Grant T32GM007276 (to D.L.). P.W.S. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- CV
coefficient of variation
- ECM
extracellular matrix
- RNA-seq
RNA sequencing
- TPM
transcripts per million
- TRN
touch receptor neuron.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0473) on September 21, 2016.
REFERENCES
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an α-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran R, Castelblanco L, Tang G, Shapiro I, Goncharov A, Jin Y. Motor neuron synapse and axon defects in a C. elegans alpha-tubulin mutant. PLoS One. 2010;5:e9655. doi: 10.1371/journal.pone.0009655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke O, Forer A. Evidence for four classes of microtubules in individual cells. J Cell Sci. 1967;2:169–192. doi: 10.1242/jcs.2.2.169. [DOI] [PubMed] [Google Scholar]
- Bounoutas A, Kratz J, Emtage L, Ma C, Nguyen KC, Chalfie M. Microtubule depolymerization in Caenorhabditis elegans touch receptor neurons reduces gene expression through a p38 MAPK pathway. Proc Natl Acad Sci USA. 2011;108:3982–3987. doi: 10.1073/pnas.1101360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, O’Hagan R, Chalfie M. The multipurpose 15-protofilament microtubules in C. elegans have specific roles in mechanosensation. Curr Biol. 2009;19:1362–1367. doi: 10.1016/j.cub.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chakraborti S, Natarajan K, Curiel J, Janke C, Liu J. The emerging role of the tubulin code: from the tubulin molecule to neuronal function and disease. Cytoskeleton. 2016. , doi: 10.1002/cm.21290. [DOI] [PubMed]
- Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Hart AC, Rankin CH, Goodman MB. Assaying mechanosensation. WormBook. 2014 doi: 10.1895/wormbook.1.172.1. 2014(Jul 31), doi: 10.1895/wormbook.1.172.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Thomson JN. Organization of neuronal microtubules in the nematode Caenorhabditis elegans. J Cell Biol. 1979;82:278–289. doi: 10.1083/jcb.82.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Thomson JN. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J Cell Biol. 1982;93:15–23. doi: 10.1083/jcb.93.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueva JG, Hsin J, Huang KC, Goodman MB. Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr Biol. 2012;22:1066–1074. doi: 10.1016/j.cub.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MP. Colchicine inhibition of nerve fiber formation in vitro. J Cell Biol. 1972;53:164–176. doi: 10.1083/jcb.53.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira HB, Zhang Y, Zhao C, Emmons SW. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Siddiqui ZK, Chou M, Culotti JG, Gogonea CB, Siddiqui SS, Hamelin M. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J Cell Sci. 1999;112:395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Yasuda H, Siddiqui SS. Molecular cloning and developmental expression of the alpha-2 tubulin gene of Caenorhabditis elegans. J Mol Biol. 1993;234:1290–1300. doi: 10.1006/jmbi.1993.1685. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Yasuda H, Siddiqui SS. Selective expression of the tba-1 α tubulin gene in a set of mechanosensory and motor neurons during the development of Caenorhabditis elegans. Biochim Biophys Acta. 1995;1261:401–416. doi: 10.1016/0167-4781(95)00028-f. [DOI] [PubMed] [Google Scholar]
- Fulton C, Simpson P. In: Cell Motility. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1976. Selective synthesis and utilization of flagellar tubulin. The multi-tubulin hypothesis; pp. 987–1005. [Google Scholar]
- Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gogonea CB, Gogonea V, Ali YM, Merz K M, Jr, Siddiqui SS. Computational prediction of the three-dimensional structures for the Caenorhabditis elegans tubulin family1. J Mol Graph Model. 1999;17:90–100. doi: 10.1016/s1093-3263(99)00025-x. [DOI] [PubMed] [Google Scholar]
- Hamelin M, Scott IM, Way JC, Culotti JG. The mec-7 beta-tubulin gene of Caenorhabditis elegans is expressed primarily in the touch receptor neurons. EMBO J. 1992;11:2885–2893. doi: 10.1002/j.1460-2075.1992.tb05357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Thein M, Brust-Mascher I, Civelekoglu-Scholey G, Lu Y, Acar S, Prevo B, Shaham S, Scholey JM. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol. 2011;13:790–798. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI. Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev Cell. 2006;10:379–390. doi: 10.1016/j.devcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Hoyle HD, Raff EC. Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol. 1990;111:1009–1026. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Miller RM, Núñez L, Portman DS. Specific α- and β-tubulin isotypes optimize the functions of sensory cilia in Caenorhabditis elegans. Genetics. 2010;185:883–896. doi: 10.1534/genetics.110.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS ONE. 2013;8:e53419. doi: 10.1371/journal.pone.0053419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux P, Steel VL, Regal C, Adgate L, Buxbaum RE, Heidemann SR. Extracellular matrix allows PC12 neurite elongation in the absence of microtubules. J Cell Biol. 1990;110:71–79. doi: 10.1083/jcb.110.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludueña RF. Are tubulin isotypes functionally significant. Mol Biol Cell. 1993;4:445–457. doi: 10.1091/mbc.4.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Manser J, Wood WB. Mutations affecting embryonic cell migrations in Caenorhabditis elegans. Dev Genet. 1990;11:49–64. doi: 10.1002/dvg.1020110107. [DOI] [PubMed] [Google Scholar]
- May GS. The highly divergent beta-tubulins of Aspergillus nidulans are functionally interchangeable. J Cell Biol. 1989;109:2267–2274. doi: 10.1083/jcb.109.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabayashi T, Palfreyman MT, Sluder AE, Slack F, Sengupta P. Expression and function of members of a divergent nuclear receptor family in Caenorhabditis elegans. Dev Biol. 1999;215:314–331. doi: 10.1006/dbio.1999.9470. [DOI] [PubMed] [Google Scholar]
- Mohri H. Amino-acid composition of “Tubulin” constituting microtubules of sperm flagella. Nature. 1968;217:1053–1054. doi: 10.1038/2171053a0. [DOI] [PubMed] [Google Scholar]
- O’Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- Ortiz CO, Etchberger JF, Posy SL, Frøkjær-Jensen C, Lockery S, Honig B, Hobert O. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006;173:131–149. doi: 10.1534/genetics.106.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics. 2015;201:47–54. doi: 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold BC, Park S-J, Mazzochette EA, Goodman MB, Pruitt BL. MEMS-based force-clamp analysis of the role of body stiffness in C. elegans touch sensation. Integr Biol. 2013;5:853–864. doi: 10.1039/c3ib20293c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Lyczak R, Ellis GC, Bowerman B. Roles for two partially redundant α-tubulins during mitosis in early Caenorhabditis elegans embryos. Cell Motil Cytoskeleton. 2004;58:112–126. doi: 10.1002/cm.20003. [DOI] [PubMed] [Google Scholar]
- Portman DS, Emmons SW. Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev Biol. 2004;270:499–512. doi: 10.1016/j.ydbio.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniello R, Arrigoni F, Bassi MT, Borgatti R. Mutations in α- and β-tubulin encoding genes: implications in brain malformations. Brain Dev. 2015;37:273–280. doi: 10.1016/j.braindev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Savage C, Hamelin M, Culotti JG, Coulson A, Albertson DG, Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3:870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- Savage C, Xue Y, Mitani S, Hall D, Zakhary R, Chalfie M. Mutations in the Caenorhabditis elegans beta-tubulin gene mec-7: effects on microtubule assembly and stability and on tubulin autoregulation. J Cell Sci. 1994;107:2165–2175. doi: 10.1242/jcs.107.8.2165. [DOI] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986;6:3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EM, Kato M, Sternberg PW. Functional transcriptomics of a migrating cell in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:16246–16251. doi: 10.1073/pnas.1203045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki H, Shinomoto S. A method for selecting the bin size of a time histogram. Neural Comput. 2007;19:1503–1527. doi: 10.1162/neco.2007.19.6.1503. [DOI] [PubMed] [Google Scholar]
- Spencer WC, McWhirter R, Miller T, Strasbourger P, Thompson O, Hillier LW, Waterston RH, Iii DMM. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PLoS One. 2014;9:e112102. doi: 10.1371/journal.pone.0112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange K, Christensen M, Morrison R. Primary culture of Caenorhabditis elegans developing embryo cells for electrophysiological, cell biological and molecular studies. Nat Protoc. 2007;2:1003–1012. doi: 10.1038/nprot.2007.143. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. The Nematode Caenorhabditis Elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. Methods; pp. 587–606. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Takeishi A, Yu YV, Hapiak VM, Bell HW, O’Leary T, Sengupta P. Receptor-type guanylyl cyclases confer thermosensory responses in C. elegans. Neuron. 2016;90:235–244. doi: 10.1016/j.neuron.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Keller C, Kalebic N, Nguyen KCQ, Somhegyi H, Politi KA, Heppenstall P, Hall DH, Chalfie M. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol. 2012;22:1057–1065. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A. Nerve regeneration in C. elegans after femtosecond laser axotomy. Nature. 2005;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci USA. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- Zheng C, Diaz-Cuadros M, Chalfie M. Hox genes promote neuronal subtype diversification through posterior induction in Caenorhabditis elegans. Neuron. 2015a;88:514–527. doi: 10.1016/j.neuron.2015.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Jin FQ, Chalfie M. Hox proteins act as transcriptional guarantors to ensure terminal differentiation. Cell Rep. 2015b;13:1343–1352. doi: 10.1016/j.celrep.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence files for raw RNA-seq reads have been deposited in the National Center for Biotechnology Information Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) under the accession numbers SRR3481679 (PLM neuron pool), SRR3481680 (AFD neuron pool), and SRR3481678 (ASER neuron pool).