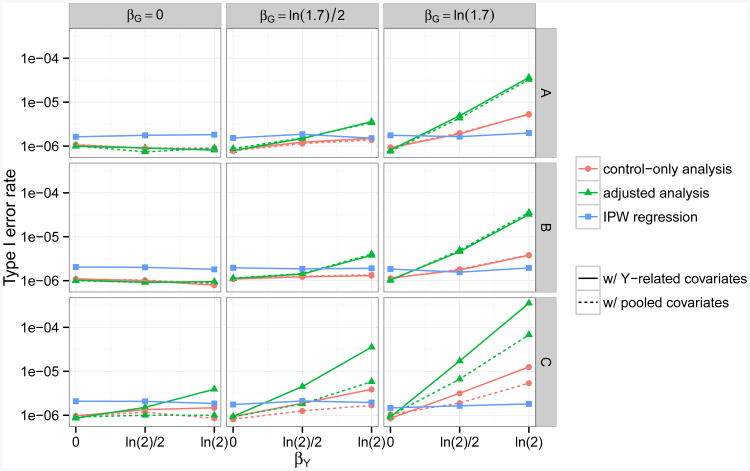
Fig 1.
Empirical type I error rates for testing genetic associations with a continuous secondary trait, at genome-wide α = 10−6 level and across scenarios with different combinations of βY, βG, γ1 and βZ1. Five methods (Analysis 2,4,6,8,9) are compared here. Each method takes either a control-only, adjusted, or IPW approach, and adjusts for covariates related to Y or covariates related to (Y, D). The disease is assumed to be common (10% prevalence) and to follow a logistic model. In row A, covariate Z1 is assumed to be associated with G but not with D (γ1 = ln 1.7, βZ1 = 0). In row B, Z1 is associated with D but not with G (γ1 = 0, βZ1 = ln 1.7). In row C, Z1 is a confounder of the association between G and D (γ1 = βZ1 = ln 1.7).