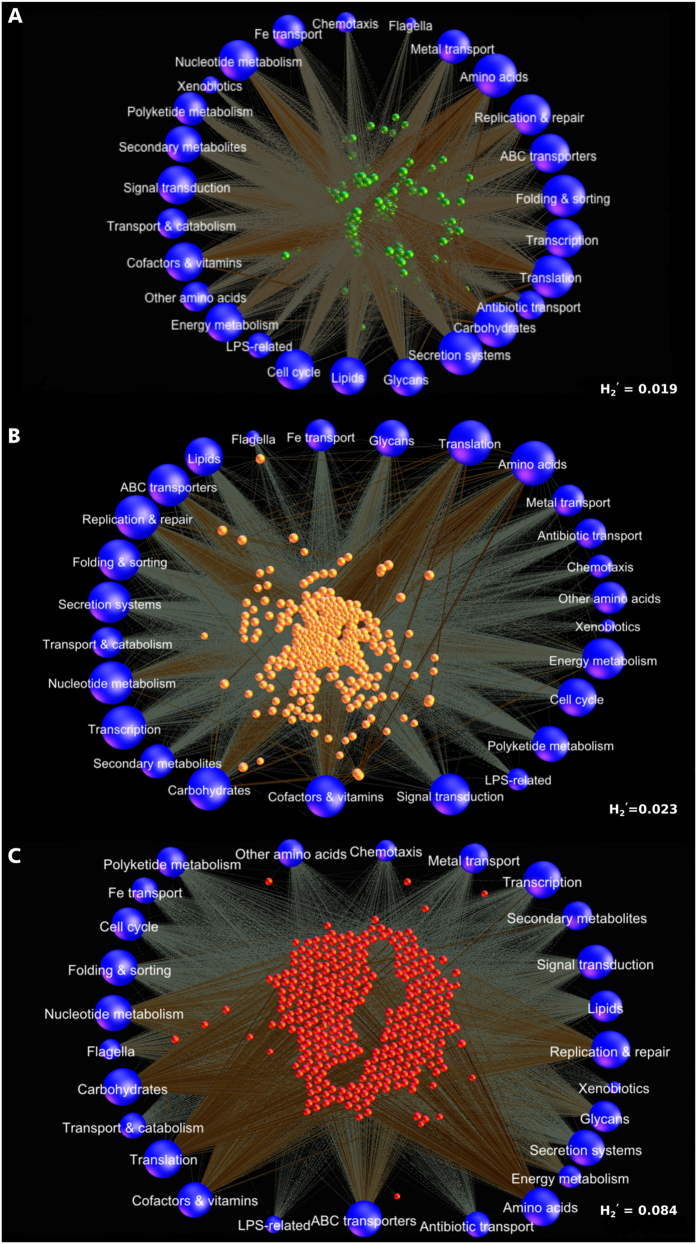
Figure 6. Functional contributions of bacterial species in the subgingival metagenome.
Force-directed networks of bacterial species and their contribution to metabolic pathways in health (A), shallow sites (B) and deep sites (C). Each network graph contains nodes (circles) and edges (lines). Nodes in the center of each network represent species-level OTU’s in healthy (green), deep sites (red) and shallow sites (orange) and nodes on the outer edge represent the functional contributions of these species. Edges represent the number of genes contributed by each species to each functional family. Only significant correlations between species and their functional contributions (p < 0.05, t-test) with a coefficient of at least 0.75 are shown. The data used to create these networks are presented in Supplementary Table 5. Few species-level nodes can be seen in health, with equal number of edges connecting each of these species to the functional nodes. Both deep and shallow sites demonstrate larger numbers of species-level nodes than health. Moreover, while many functions species are connected to their cognate species by large numbers of edges, certain functions have contributions only from a few species. This is numerically indicated by the degree of functional specialization (H2′).