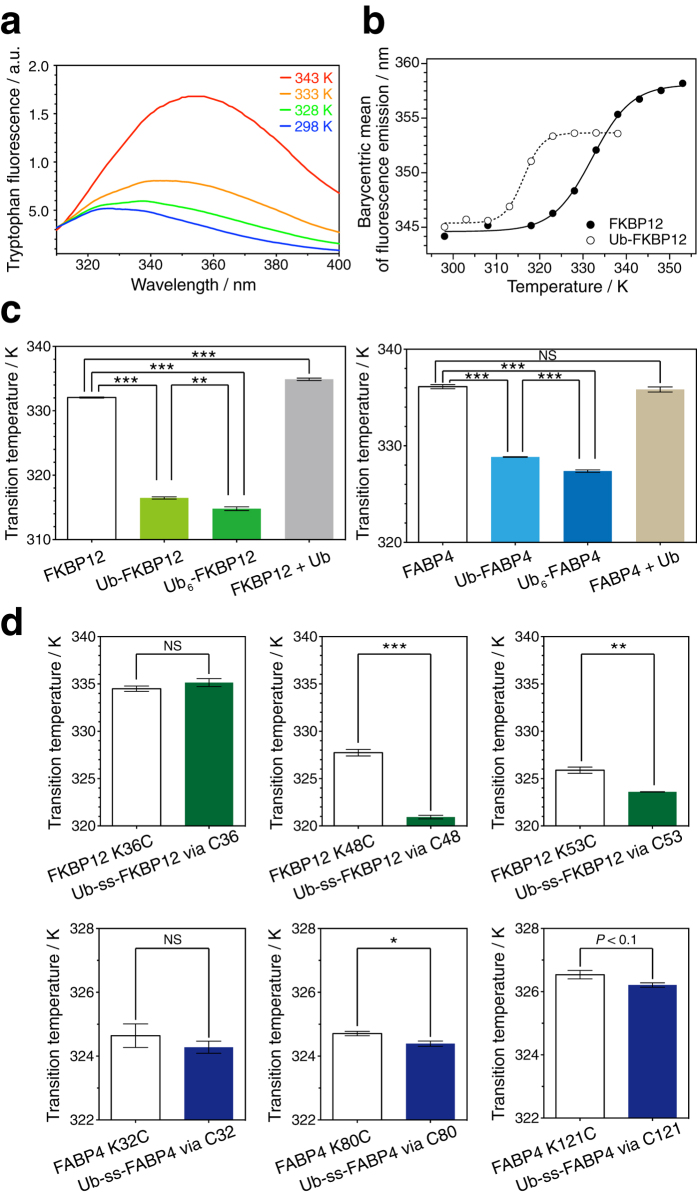
Figure 2. Fold destabilization of a substrate proteins by (poly-)ubiquitylation.
(a) Temperature-dependent changes in the tryptophan fluorescence emission spectra of FKBP12. The emission maximum in the fluorescence spectra shows a red shift with increasing temperature. (b) Series of fluorescence emission wavelengths (barycentric mean) of FKBP12 (filled circles) and N-terminally mono-ubiquitylated FKBP12 (open circles). The data were fitted to the sigmoidal equation (solid and dashed line, respectively). (c) Comparative analysis of the thermal denaturation transition for N-terminal (poly-)ubiquitylated FKBP12 (left) and FABP4 (right). Simple addition of ubiquitin to a substrate protein had little effect on its thermostability. (d) Comparative analysis of thermal denaturation transitions for chemically ubiquitylated FKBP12 (upper) and FABP4 (lower). For disulfide conjugation, FKBP12 and FABP4 carry the additional mutations C22S and C2A/C118A, respectively. ss indicates the disulfide bridge. All values represent the average of three independent experiments. Error bars indicate the standard error of the mean. ***P < 0.001, **P < 0.01, and *P < 0.05 (Student’s t test). NS indicates no statistical significance: P > 0.1. Transition temperature values and statistics are shown in Table 1.