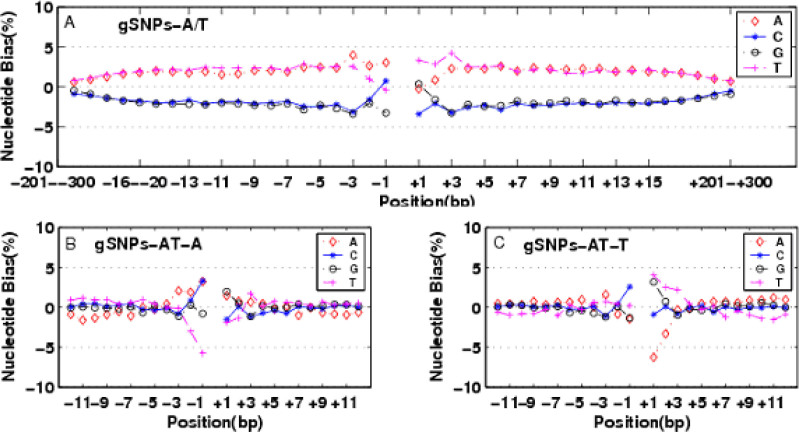
Fig. 2.
The impact of expected nucleotide proportion for NNEs. A. gSNPs-A/T. The expected nucleotide proportion was averaged over the whole rice genome. The PLUS-bias of AT nucleotides extended nearly 300 bp to both sides in gSNPs-A/T; B. gSNPs-A/T-A; C. gSNPs-A/T-T. The expected nucleotide proportion was calculated based on the flanking sequence of the corresponding categories. The long effect range of NNEs disappeared and it was only limited 3 bp to SNP sites in gSNPs-C/T-C and gSNPs-A/T-T.