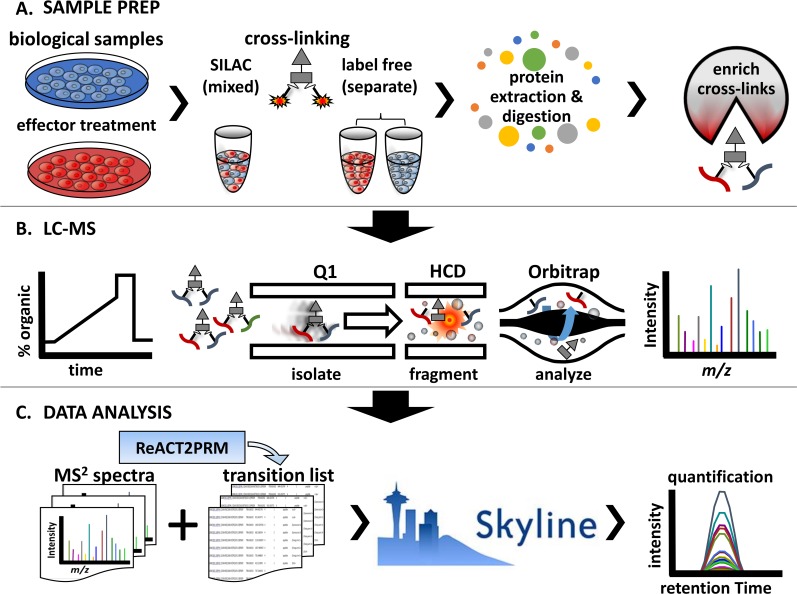
Fig 1. Experimental outline.
A. Biological samples are prepared for qXL-MS comparing two or more conditions. The samples are treated with chemical cross-linker either as (1) a mixed sample if SILAC labeling was used or (2) as separate samples if carrying out a label free experiment or using isotopically labeled cross-linkers. Following the cross-linking reaction proteins are extracted, enzymatically digested, and subjected to various strategies (i.e. strong cation exchange and affinity chromatography) for enrichment cross-linked peptide pairs. B. LC-MS analysis of samples enriched for cross-linked peptide pairs is carried out. This consists of reversed phase chromatographic separation by LC followed by analysis by MS. The mass spectrometer is operated in PRM mode where an inclusion list of m/z values for the precursor ions of interest is used to target specific cross-linked peptides. The PRM mass spectrometric analysis used here consists of three steps including isolation of precursor ions, fragmentation by collision with neutral gasses, and detection of mass to charge ratios of the resulting fragment ions. C) Resulting MS2 data are converted into transition lists and imported into Skyline for analysis.