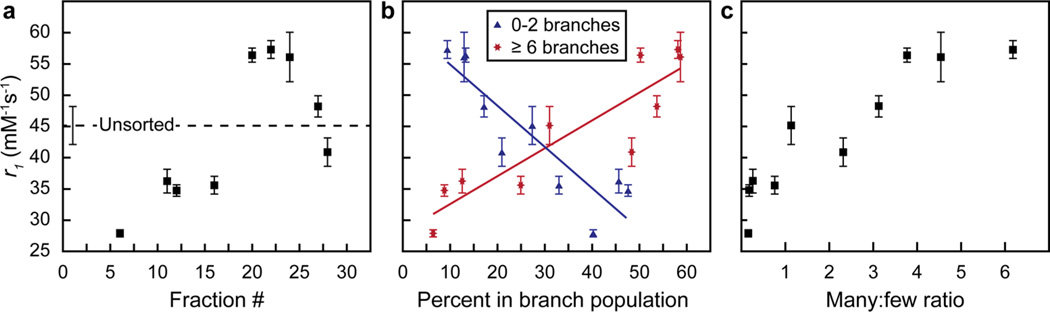
Figure 3.
Distribution of many- and few-branched gold nanostars influences the relaxivity of Gd(III)-DNA. r1, measured at 37 °C and 1.41 T (60 MHz), (a) for each fraction, (b) as a function of the percentage of many- or few-branched DNA-Gd@stars (solid lines: linear fit), and (c) as a function of the ratio of many:few-branched particles. Error bars are the standard deviation from two r1 measurements of each sample.