Figure 5. GAPLINC bound to PSF and NONO.
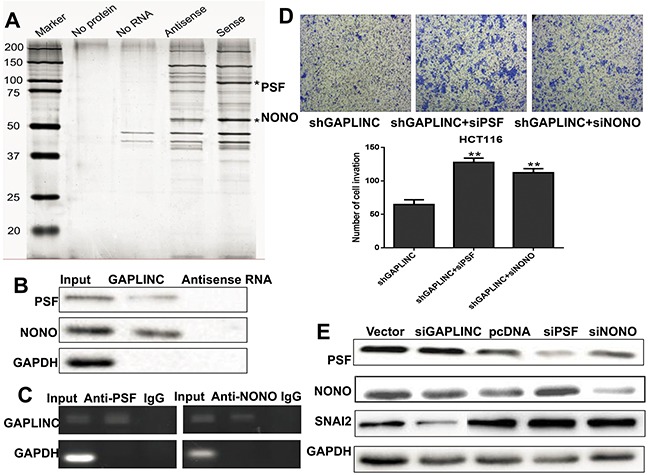
A. RNA pull-down was performed using a GAPLINC template and RNA-bound protein separated by SDS-PAGE in HCT116. The protein bands were excised and detected by mass spectrometry analysis. B. PSF and NONO were detected by the Western blotting assay in the samples pulled down by GAPLINC. C. RIP analyses were performed using antibodies against PSF and NONO, with IgG as a negative control in HCT116. The enrichment of the GAPLINC was detected using RT-PCR and normalized to the input. D. The Transwell assay was performed to assess the invasion ability of HCT116-GAPLINC-shRNA cells transiently transfected with empty vector, PSF-siRNA, and NONO-siRNA. E. Western blotting analysis showed that the protein levels of PSF, NONO, SNAI2, and GAPDH were detected in HCT116 empty vector, GAPLINC-siRNA, GAPLINC-pcDNA3.1, PSF-siRNA, and NONO-siRNA cells. Data represent mean ± standard deviation from three independent experiments. *P < 0.05, **P < 0.01.
