Abstract
To determine the mechanism by which fibroblast growth factor 9 (FGF9) alters granulosa (GC) and theca (TC) cell proliferation, cell cycle proteins that regulate progression through G1 phase of the cell cycle, cyclin D1 (CCND1) and cyclin-dependent kinase-4 (CDK4; CCND1's catalytic partner), were evaluated. Ovaries were obtained from a local abattoir, GC were harvested from small (1-5 mm) and large (8-22 mm) follicles, and TC were harvested from large follicles. GC and TC were plated in medium containing 10% fetal calf serum followed by various treatments in serum-free medium. Treatment with 30 ng/mL of either FGF9 or IGF1 significantly increased GC numbers and when combined, synergized to further increase GC numbers by threefold. Abundance of CCND1 and CDK4 mRNA in TC and GC were quantified via real-time PCR. Alone and in combination with IGF1, FGF9 significantly increased CCND1 mRNA expression in both GC and TC. Western blotting revealed that CCND1 protein levels were increased by FGF9 in TC after 6 h and 12 h of treatment, but CDK4 protein was not affected. A mitogen-activated protein kinase (MAPK)/ extracellular signal-regulated kinase (ERK) pathway inhibitor, U0126, significantly reduced FGF9-induced CCND1 mRNA expression to basal levels. For the first time we show that CCND1 mRNA expression is increased by FGF9 in bovine TC and GC, and that FGF9 likely uses the MAPK pathway to induce CCND1 mRNA production in bovine TC.
Keywords: Fibroblast growth factor 9 (FGF9), cyclin-dependent kinase 4 (CDK4), cyclin D1 (CCND1), granulosa cells, theca cells, cattle
1. Introduction
The fibroblast growth factor (FGF) family consists of 22 polypeptides that act primarily as paracrine signaling molecules and bind to one or more of the FGF receptors (FGFRs) that include: FGFR1b, FGFR1c, FGFR2b, FGFR2c, FGFR3c, and FGFR4 (Chaves et al., 2012; Laestander and Engström, 2014). Members of the FGF family are further divided into seven sub-families based on their receptor affinities; these families include FGF1 (includes FGF1 which binds all FGF receptors and FGF2 which binds to FGFR1c and −2c), FGF4 (includes FGF4, −5, and −6 which binds to FGFR1c and −2c), FGF7 (includes FGF7, −3, −10, and −22 which bind to FGFR2b and −1b), FGF8 (includes FGF8, −17, and −18 which bind to FGFR3c, 4, and 1c), FGF9 (includes FGF9, −16, and −20 which primarily bind to FGFR3c, −2c, and −4), FGF19 (includes FGF19, −21, and −23; endocrine factors that weakly bind to FGFR1c and −2c), and FGF11 (includes FGF11, −12, −13, and −14 which do not activate FGFRs) (Ornitz et al., 1996; Pownall and Isaacs, 2010; Laestander and Engström, 2014). However, not all FGFs or FGFRs are present in every cell type of every species.
In ovarian tissue of cattle, FGF2 is localized to granulosa cells (GC) and theca cells (TC) (van Wezel et al., 1995), FGF7 is present in TC (Parrott and Skinner, 1998), FGF8, −9, −10, and −17 are expressed in both GC and TC (Buratini et al., 2005, 2007; Drummond et al., 2007; Machado et al., 2009; Schreiber et al., 2012), and FGF18 is present in TC (Portela et al., 2010). Furthermore, FGFR2b, −2c, −3b and −3c are expressed in bovine GC (Parrott and Skinner, 1998; Berisha et al. 2000, 2004; Buratini et al. 2005), while FGFR2c, −3c and −4 are expressed in bovine TC (Berisha et al., 2004; Buratini et al., 2005). Thus, it should be expected that GC and TC would respond differently to the various FGFs.
FGF9 gene expression was first discovered in the rat ovary by Drummond et al. (2007) and later in the bovine ovary by Grado-Ahuir et al. (2011) who found that FGF9 mRNA in GC was down-regulated in ovarian follicular cysts compared to normal-follicles. Subsequent studies found that FGF9 treatment significantly increases bovine GC and TC proliferation while inhibiting steroidogenesis in vitro (Schreiber and Spicer, 2012; Schreiber et al., 2012). Furthermore, FGF9 mRNA is present in both TC and GC, with cells from small follicles having greater relative abundance compared to cells from large follicles (Schreiber and Spicer, 2012; Schreiber et al., 2012), and GC having a greater abundance of FGF9 mRNA than TC (Schreiber et al., 2012) indicating that FGF9 may be playing both a paracrine and an autocrine role in growing follicles.
An increase in cell proliferation is mediated by increased speed of the cell cycle which is divided into four phases: gap 1 (G1), synthesis (S), gap 2 (G2), and mitosis (M) (Lim and Kaldis, 2013). Each of these phases are regulated by different proteins at specific checkpoints, insuring that the cell cycle progresses in the correct order, and only when the cell is healthy. Checkpoints include the restriction (R) point within G1, G1/S, S/G2, G2/M and M (Nigg, 2001; Lim and Kaldis, 2013). The most important protein families for cell cycle regulation consist of cyclins (CCNs), cyclin-dependant kinases (CDKs), and cyclin-dependant kinase inhibitors (CDKIs) (Bendris et al., 2015). Cyclins are the regulatory subunit of CDKs and once they are bound together, the dimer can activate its respective checkpoint to allow progression of the cell cycle; however, dimers can also be inactivated by CDKIs (Bendris et al., 2015). Each checkpoint is activated by specific proteins as follows: R point, CCND and CDK4 or −6; G1/S phase, CCNE and CDK2; S/G2 phase, CCNA and CDK2; G2/M phase, CCNA and CDK1; and M phase, CCNE and CDK1 (Nigg, 2001; Alberts et al., 2002; Lim and Kaldis, 2013). Therefore, we hypothesized that FGF9 increases cell proliferation by altering specific cell cycle proteins. Previous studies with ovarian endometroid adenocarcinomas (Schwartz et al., 2003), human uterine endometrial stromal cells (Wing et al., 2005) and endometrial cancer tissues (Chan et al., 2012) have shown a link between FGF9 and CCND1. Therefore, our objectives were to characterize the change in CCND1 and CDK4 mRNA after FGF9 treatment of bovine GC and TC, and to determine which intracellular pathway mediated this change.
2. Materials and Methods
2.1 Reagents and Hormones
Reagents used during cell culture included: Ham's F-12, Dulbecco modified Eagle medium (DMEM), gentamicin, sodium bicarbonate, streptomycin/penicillin, TRI reagent, trypan blue, protease, collagenase, hyaluronidase, and deoxyribonuclease from Sigma-Aldrich Chemical Company (St. Louis, MO); and fetal calf serum (FCS) from Atlanta Biologicals (Atlanta, GA).
The hormones and inhibitors used during cell culture included: ovine FSH (175 × NIH-FSH-S1 U/mg) and ovine LH (NIADDK-NIH-26; AFP5551B) from the National Hormone and Peptide Program (Harbor-UCLA Medical Center, Torrance, CA); testosterone from Steraloids (Newport, RI); recombinant human IGF1 and FGF9 (without carrier protein) from R&D Systems, Inc. (Minneapolis, MN); LY294002 (a PI3K inhibitor), H89 (a PKA inhibitor), U0126 (a MAPK/ERK inhibitor) and wortmannin (a PI3K inhibitor) from Enzo Life Sciences Inc. (Farmingdale, NY).
2.2 Cell Culture
Ovaries were collected from cattle at a local abattoir, and follicular fluid was aspirated from small (1 to 5 mm) and large (8 to 22 mm) follicles, and GC and TC were isolated as previously described (Langhout et al., 1991; Lagaly et al., 2008; Spicer et al., 2002; 2009). Isolated GC and TC were resuspended in medium containing 1.5 mg/mL of collagenase and 0.5 mg/mL of DNase to prevent clumping as previously described (Lagaly et al., 2008; Spicer et al., 2009). Cell viability was determined by trypan blue exclusion method on a 0.1 mm deep hemocytometer (American Optical Corporation, Buffalo, NY). Viability of bovine GC from small and large follicles and TC from large follicles averaged 69%, 61%, and 92%, respectively.
Cells were plated on 24-well Falcon multi-well plates (Becton Dickinson, Franklin Lakes, NJ) in 1 mL of medium (1:1 DMEM and Ham's F12 with 2.0 mM glutamine, 0.12 mM gentamicin, and 38.5 mM sodium bicarbonate) or in 60 mm Falcon culture dishes (Becton Dickson) in 4 mL of medium with and average plating density of 2.0 × 105 (range 0.8 to 4.4 × 105) or 8.8 × 105 viable cells per well or plate, respectively, and cultured at 38.5°C with 5% CO2. Cells were cultured in the presence of 10% FCS until they reached 80% confluency (i.e., 48 h to 96 h) with a medium change every 24 h. Cells were then washed twice with 0.5 mL of serum-free medium followed by treatment with various hormones, administered in serum-free medium, for 6 h, 12 h, 24 h, and/or 48 h depending on the experiment.
2.3 Experimental Design
Experiment 1 was conducted to evaluate the interaction between FGF9 and IGF1 on cell proliferation. Granulosa cells from small- and large-follicle were cultured for 48 h in 10% FCS, and then GC were washed with serum-free medium and treated with 30 ng/mL of FSH and 30 ng/mL of IGF1, 30 ng/mL of FGF9 or both for an additional 48 h. GC were treated with FSH because the cell proliferation responses to insulin, IGF1 and FGF9 are greater in its presence than its absence (Spicer et al., 2002; Spicer and Aad, 2007; Schreiber and Spicer, 2012).
Experiment 2 was designed evaluate if FSH and IGF1 interact with FGF9 to regulate CCND1 mRNA accumulation in small-follicle GC. After 48 h of culture in 10% FCS, GC were washed in serum-free medium and serum-starved for 24 h in order to synchronize cells in G1 of the cell cycle (Wing et al., 2005). Cells were then treated with either no addition (control), FSH (30 ng/ml) or IGF1 (30 ng/ml) with or without 30 ng/mL of FGF9 for 12 h and RNA was collected.
Experiment 3 was conducted to evaluate the effects of FGF9 on CCND1 and CDK4 gene expression in small- and large-follicle GC. After 48 h of culture in 10% FCS, GC were washed with serum-free medium and treated with 30 ng/mL of FSH and 30 ng/mL of IGF1 with or without 30 ng/mL of FGF9 and RNA was collected 24 h after treatment. GC were treated with both FSH and IGF1 because the cell proliferation response to FGF9 is greater in their presence than their absence (Schreiber and Spicer, 2012).
Experiment 4 was designed to determine the effect of LH on the CCND1 and CDK4 mRNA response to FGF9. After 72 h in 10% FCS medium, large-follicle TC were washed in serum-free medium and treated with IGF1 (30 ng/mL) and either no other additions (Control), LH (30 ng/mL), FGF9 (30 ng/mL), or both LH and FGF9; RNA was collected 12 h after treatment.
Experiment 5 was designed to determine which intracellular pathway(s) FGF9 uses to increase CCND1 and/or CDK4 mRNA expression. After 72 h in 10% FCS, large-follicle TC were washed with serum-free medium and treated with: 1) vehicle control; 2) 30 ng/mL of IGF1 and 30 ng/mL of FGF9; 3) FGF9 plus IGF1 and 20 M of U0126, a MAPK/ERK inhibitor; 4) FGF9 plus IGF1 and 10 M of H89, a PKA inhibitor; 5) FGF9 plus IGF1 and 200 nM of wortmannin, a PI3K/AKT inhibitor; or 6) FGF9 plus IGF1 and 10 M of LY294002, a PI3K/AKT inhibitor. RNA was collected 24 h after treatment. The selected doses of inhibitors were based on previous studies (Asselin et al., 2001; Poretsky et al., 2001; Dewi et al., 2002; Laurich et al., 2002; Silva et al., 2006).
Experiment 6 was designed to determine the effect of U0126 on CCND1 mRNA expression in IGF1- and FGF9-treated large-follicle TC. After 96 h in 10% FCS medium, large-follicle TC were washed in serum-free medium and treated with no addition (Control), IGF1 (30 ng/mL) or FGF9 (30 ng/ml) with or without 20 μM of U0126. RNA was collected 24 h after treatment.
Experiment 7 was designed to determine the effect of FGF9 on CCND1 and CDK4 protein accumulation in large-follicle TC. TC were cultured in either 24-well culture plates or 60 mm dishes with medium containing 10% FCS media for 96 h, then washed in serum-free medium, and serum-starved for 24 h in order to synchronize cells in G1 of the cell cycle (Wing et al., 2005). Cells were then treated with or without 30 ng/mL of FGF9 and cellular protein was collected 0 h, 6 h, and 12 h after treatment.
2.4 RNA Extraction and Real-time PCR
At the end of the treatment period, medium was aspirated and cells from 2 replicate wells of a 24-well plate were lysed in 0.5 mL of TRI Reagent and RNA isolated as previously described (Lagaly et al., 2008; Grado-Ahuir et al., 2011). RNA was quantitated with Nano-Drop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE) at 260 nm, and diluted to 10 ng/μL in DEPC-treated water and stored at −80°C.
For quantitative PCR, CCND1 and CDK4 primers and probes were designed using Primer Express™ Software (Applied Bio Systems, Foster City, CA). The CCND1 (1199 base pairs (bp), Accession NM_001046273) forward primer [CGACTTCATCGAGCACTTCCT] was between bp 634 and 654, with annealing Tm of 57°C. The CCND1 reverse primer [TCTGTGCCACAGACGTGAAGTT] was between bp 723 and 744, with an annealing Tm of 59°C. The CCND1 probe [ATGCCGGTGGCCGAGGAGAAC] was between bp 662 and 682, with annealing Tm of 65°C and contains 5’ reporter dye FAM and 3’ quencher dye TAMRA. The CDK4 (1369 bp, Accession NM_001037594) forward primer [ACTCTGGTATCGTGCTCCAGAAG] was between bp 671 and 693, with an annealing Tm of 58°C. The CDK4 reverse primer [CAGAAGAGAGGCTTTCGACGAA] was between bp 784 and 763, with an annealing Tm of 57°C. The CDK4 probe [CGTATGCAAC ACCCGTGGACATGTG] was between bp 709 and 733, with an annealing Tm of 63°C and contains 5’ reporter dye FAM and 3’ quencher dye TAMRA. After RNA extraction and dilution, 50 ng of RNA samples were loaded into a clear 96-well PCR plate (BioRad Laboratories, Hercules, CA) in duplicate design with both target genes, CDK4 and CCND1, as well as the housekeeping gene, 18S, on a single plate. Single-plex master mixes for target genesCDK4 and CCND1 were composed of: 400 nM forward primer, 400 nM reverse primer, 200 nM fluorescent-labeled (FAM) probe, reaction mix, mScribe, and DEPC-treated water. Single-plex master mixes for Ribosomal 18S RNA Control (Life Technologies, Inc., Grand Island, NY) were composed of: 100 nM of each forward primer, reverse primer, and fluorescent-labeled (VIC) probe, reaction mix, mScribe, and DEPC-treated water. Plates were then sealed with caps (BioRad Laboratories) and centrifuged in MPS1000 Mini PCR Plate Spinner (Labnet, Edison, NJ) for 15 sec and placed in a CFX96 Touch Real Time PCR C1000 Thermal Cycler (BioRad Laboratories) set to parameters, as described previously (Aad et al., 2006; Grado-Ahuir et al., 2011). For all PCR runs, no template controls were included to insure the lack of contaminants in the master mix and the absence of any genomic DNA contamination, respectively. Data were evaluated using the comparative threshold cycle (CT) method as previously described (Voge et al., 2004; Aad et al., 2006; Grado-Ahuir et al., 2011). The CV for the Ct of CCND1, CDK4 and 18S mRNA duplicates averaged 1.5 ± 0.4%, 1.4 ± 0.4%, and 1.3 ± 0.2%, respectively.
2.6 Protein Isolation
At the end of the treatment period, medium was aspirated, cells were washed twice with 0.5 mL (24-well plates) or 2 mL (60 mm dishes) of PBS and lysed with either 0.2 mL (24-well plates) or 0.15 mL (60 mm dishes) Mammalian Protein Extraction Reagent (MPER; Thermo Fisher Scientific, Inc., Rockford, IL) containing 1% EDTA-Free Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific) and scraped with a cell scraper (BD Bioscience Discovery Labware, Bedford, MA). After MPER treatment, 2 wells were pooled (24-well plates) or a sample from a single 60 mm dish were pipetted into 1.5 mL microcentrifuge tubes and stored at −80°C. Cell lysates were thawed and centrifuged at 15,000 × g for 15 min, the supernatant was transferred to a new microcentrifuge tube and cell debris pellets were discarded. Protein concentration was then quantified by Pierce BCA Assay (Pierce Biotechnology, Rockford, IL) using the SpectraMax M3 plate reader (Molecular Devices, LLC, Sunnyvale, CA) at 540 nm.
In experiments using 24-well culture plates and one pool of cells using 60 mm plates (Experiment 6), samples were determined to be too dilute for Western blot use. Samples from 24-well plates were pooled together yielding a total sample volume of 0.8 mL, then concentrated approximately 10-fold with Centricon® YM-10 microcentrifuge filters (24-well plates) or YM-3 filters (60 mm dishes) (EMD Millipore Corp., Billerica, MA). Briefly, filter devices were assembled, samples were vortexed, added to the filter reservoir, and centrifuged at 3045 × g for up to 1 h, checking every 15 min until approximately 60% of sample volume was in filtrate vial for YM-10 filters. For YM-3 filters sample was centrifuged until 90% of sample volume was in the filtrate vial. After filtration, the retentate vial was attached to the filter device, flipped up-side-down, and centrifuged at 630 x g for 5 min. After centrifugation samples were transferred to new microcentrifuge tubes and protein content was quantified again, using the Pierce BCA Assay. Sample concentration was increased an average of 9.8-fold and 7.7-fold with Centricon YM-10 and YM-3 filters, respectively.
2.7 Western Blotting
The same amount of total protein, 25 μg for CDK4 detection and 35 g for CCND1 detection, and PBS was added when needed to yield the same volume for each sample. Due to low total protein content, samples from 24-well culture plates and samples from 60-mm culture dishes (Experiment 7) were combined yielding 3 samples each from control and FGF9-treated TC. Loading dye (tris-hydrochloride buffer, bromophenol blue, SDS, β–mercaptoethanol, and glycerol) was added to normalized samples at 1:3, boiled for 5 min, centrifuged for 15 sec, and loaded into 4% stacking, 12% resolving SDS-PAGE mini gels; Precision Plus Protein™ All Blue Standards (Bio-Rad, Hercules, CA) and recombinant human CDK4 (ProSpec-Tany TechnoGene Ltd., Ness Ziona, Israel) for CDK4 and Precision Plus Protein™ Kaleidoscope™ Standards (Bio-Rad Laboratories) and recombinant human CCND1 (GeneCopoeia, Rockville, MD) for CCND1 were also loaded into the gels. Gels were ran in a Mini-PROTEAN® 3 Cell Apparatus (Bio-Rad) at 200 V for 45 min, or until the dye front reached the bottom of the gel. The gels were then removed and placed in nitrocellulose transfer buffer and proteins transferred to nitrocellulose membranes with an iBlot® Gel Transfer Device (Life Technologies, Inc.). After transfer, membranes were blocked as described previously (Castanon et al., 2012) and incubated with either CDK4 antibody (rabbit polyclonal antibody against human CDK4; GeneTex, Inc., Irvine, CA) at 1:1,000 for 3 days or CCND1 antibody (rabbit polyclonal antibody against human CCND1; Santa Cruz Biotechnology, Inc., Dallas,TX) at 1:500 or 1:375 overnight at 4°C on a rocking platform shaker. Membranes were then washed and the secondary goat anti-rabbit HRP-conjugated antibody (ImmunoPure® HRP-conjugated Goat Anti-Rabbit IgG; Pierce Biotechnology, Rockford, IL; diluted in 5% milk at 1:15,000 for CDK4 detection or at 1:20,000 to 1:25,000 for CCND1 detection) was incubated with membranes for 1 h at 25°C on a rocking platform shaker after which membranes were washed. CDK4 membranes were then dried and incubated with Immobilon® Chemiluminescent HRP Substrate for 3 min, wrapped in plastic, exposed to CL-XPosure Film for 1 sec up to 10 min, and developed with a Mini-Medical X-Ray film processor (AFP Imaging, Elmsford, NY) as previously described (Castanon et al., 2012; Stapp et al., 2014). CCND1 membranes were dried incubated with SuperSignal® West Femto Maximum Sensitivity Substrate for 5 min, placed in a plastic page protector, and developed with the FluorChem™ E FE0622 Imager from ProteinSimple (San Jose, CA) on movie mode for 180 min chemiluminescence detection and automatic marker detection. After developing, membranes were washed and re-probed for β-actin (that served as a loading control; Castanon et al., 2012; Stapp et al., 2014) using rabbit monoclonal antibody against human β-actin from Cell Signaling Technology, Inc. (Danvers, MA) at concentrations of 1:10,000 to 1:15,000 for CDK4 and CCND1 target membranes overnight at 4°C on a rocking platform shaker. The membranes were then washed and incubated with secondary antibody at 1:5,000 for CDK4 membranes or 1:25,000 to 1:40,000 for CCND1 membranes on a rocking platform shaker for 1 h at 25°C. After CDK4 and corresponding β-actin bands were visualized, X-ray images were converted to black and white, background minimized, and bands sharpened with iPhoto version 8.1.2 (Cupertino, CA). Image analysis was performed with ImageJ 1.48v (NIH, Bethesda, MD). The band intensity values of CCND1 and its corresponding β-actin were calculated by the FluorChem™ Imager software. Relative β-actin values for each band were determined by dividing each β-actin density/intensity by the largest β-actin density/intensity. The amount of CDK4 and CCND1 protein for each sample was then normalized by dividing each CDK4 and CCND1 band density/intensity by its corresponding relative β-actin.
2.8 Cell counting
To determine cell numbers, culture medium was aspirated from wells, cells were washed, trypsinized and counted on a Coulter counter (Z2 Coulter® Particle Count and Size Analyzer; Beckman Coulter, Hialeah, FL) as previously described (Lagaly et al. 2008; Schreiber et al., 2012).
2.9 Statistical Analysis
Within each experiment, three independent pools of cells were used for each cell type representing experimental replicates. Each pool of small-follicle GC were obtained from 25 to 30 ovaries yielding 6 to 8 mL of follicular fluid. Each pool of large-follicle GC and TC were obtained from 5 to 7 follicles from at least three animals. For RNA experiments, medium was administered to 4 wells; duplicate samples for each pool of cells were obtained by combining samples from 2 wells. For protein samples, each experimental replicate was generated either from a single 60 mm dish, or from 4 wells in a 24-well plate combined. The treatment effects of the dependent variables (i.e., CCND1 and CDK4 mRNA abundance and protein band intensity) were identified using the general linear models (GLM) procedure of SAS for Windows (version 9.3, SAS Institute, Inc., Cary, NY). Mean differences were determined by Fisher's protected least significant differences test (Ott, 1977) if significant main effects in the ANOVA were detected. Data are presented as least square means ± SEM.
3. Results
Experiment 1
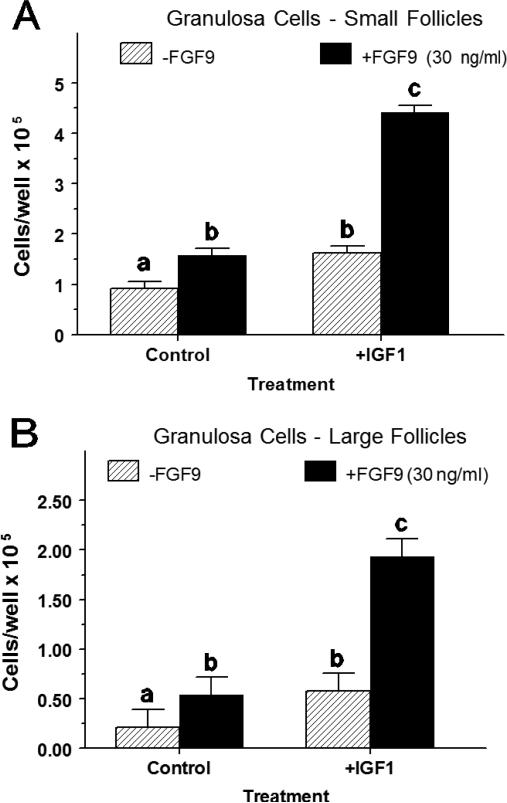
In small-follicle GC treated with FSH, IGF1 alone and FGF9 alone increased (p < 0.01) cell numbers by 1.76-fold and 1.71-fold, respectively (Figure 1A). Combined treatment of IGF1 and FGF9 increased (p < 0.05) cell numbers 2.73- to 2.82–fold above either treatment alone (Figure 1A). Similarly, in large-follicle GC treated with FSH, IGF1 alone and FGF9 alone increased (p < 0.01) cell numbers by 2.8-fold and 2.6-fold, respectively (Figure 1B). Combined treatment of IGF1 and FGF9 increased (p < 0.01) cell numbers 3.3- to 3.6–fold above either treatment alone (Figure 1B).
Fig. 1.
Interaction between IGF1 and FGF9 on cell proliferation of small- and large-follicle GC (Exp. 1). GC were cultured for 48 h as described in Materials and Methods, and then treated for an additional 48 h with FSH (30 ng/ml) and IGF1 (30 ng/ml), FGF9 (30 ng/ml) or both IGF1 and FGF9. a,b,cMeans (± SEM of three separate experiments) without a common letter differ (p < 0.05).
Experiment 2
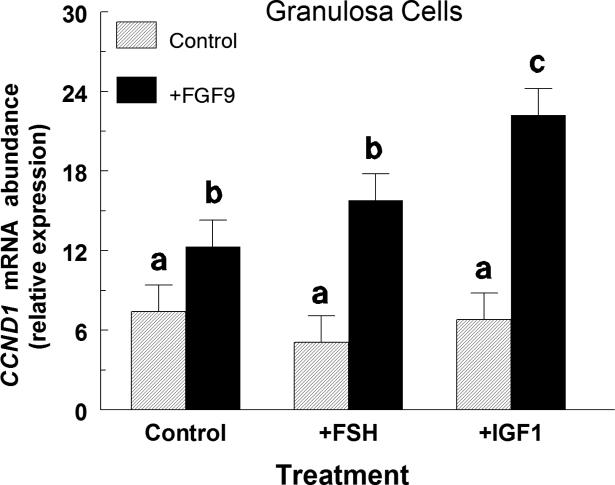
In small-follicle GC, neither FSH nor IGF1 treatment alone altered (p > 0.10) CCND1 mRNA abundance (Figure 2). However, IGF1 but not FSH significantly amplified the FGF9-stimulated CCND1 mRNA such that in the absence of IGF1, FGF9 increased (p < 0.05) CCND1 mRNA by 1.7-fold whereas in the presence of IGF1, FGF9 increased (p < 0.05) CCND1 mRNA by 3.3-fold (Figure 2). The average CCND1 Ct was 24.3 ± 0.1.
Fig. 2.
Effect of FSH, IGF1 and FGF9 on CCND1 mRNA expression in small-follicle GC (Exp. 2). Cells were cultured for 48 h as described in Materials and Methods, serum-starved for 24 h and then treated for an additional 12 h with no additions (Control), FSH (30 ng/ml) or IGF1 (30 ng/ml) and either control or FGF9 (30 ng/ml). Values are normalized to constitutively expressed 18S ribosomal RNA and are means ± SEM of three separate experiments. a,b,cMeans without a common letter differ (p < 0.05).
Experiment 3
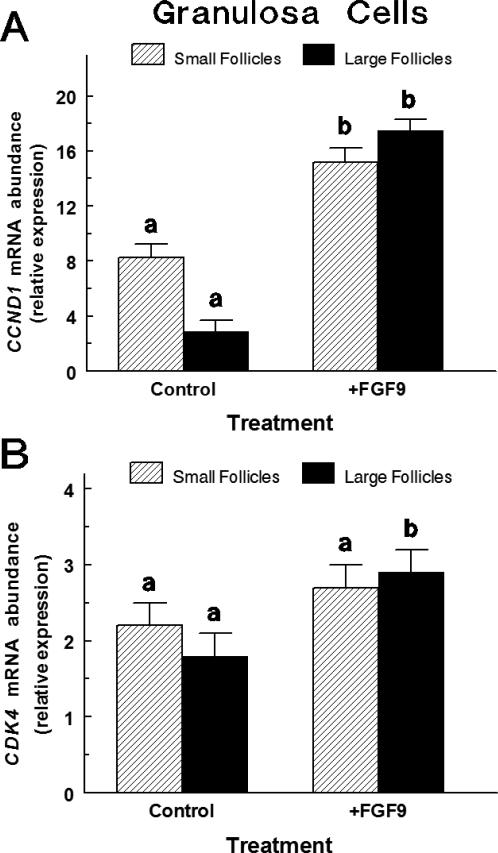
In small-follicle GC treated with FSH and IGF1, FGF9 treatment increased (p < 0.01) CCND1 mRNA abundance by 1.85-fold (Figure 3A) and tended to increase (p < 0.10) CDK4 mRNA abundance (Figure 3B). In large-follicle GC treated with FSH and IGF1, FGF9 treatment caused a 6-fold increase (p < 0.001) in CCND1 mRNA abundance (Figure 3A) and increased (p < 0.05) CDK4 mRNA abundance by 60% (Figure 3B). The CCND1 Ct and CDK4 Ct averaged 31.1 ± 0.2 and 31.4 ± 0.2, respectively.
Fig. 3.
Effect of FGF9 on CCND1 (Panel A) and CDK4 (Panel B) mRNA in small and large follicle GC (Exp. 3). Cells were cultured for 48 h as described in Materials and Methods, and then treated for an additional 24 h with either Control [IGF1 (30 ng/ml) plus FSH (30 ng/mL)] or FGF9 treatment [FGF9 (30 ng/ml) plus IGF1 plus FSH]. Values are normalized to constitutively expressed 18S ribosomal RNA and are means ± SEM of three separate experiments for large and small follicles. a,bWithin follicle size group, means without a common letter differ (p < 0.05).
Experiment 4
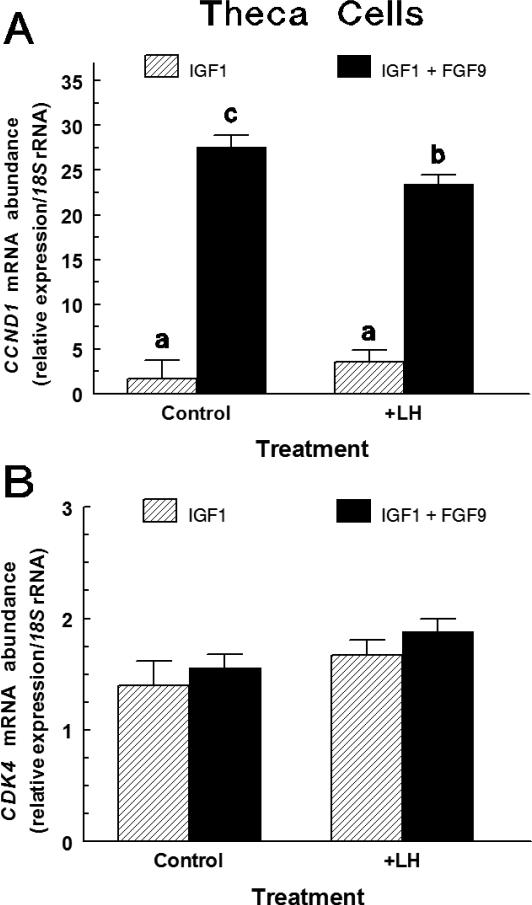
In large-follicle TC treated with IGF1, FGF9 treatment increased (p < 0.0001) CCND1 mRNA abundance by 16.2-fold (Figure 4A). A LH by FGF9 interaction tended (P < 0.06) to affect CCND1 mRNA abundance, such that co-treatment with LH suppressed (p < 0.05) the FGF9-induced increase of CCND1 mRNA with FGF9 increasing (p < 0.05) CCND1 mRNA by 6.5-fold. Neither LH nor FGF9 treatment significantly affected CDK4 mRNA expression in IGF1-treated TC (Figure 4B). The CCND1 Ct and CDK4 Ct averaged 24.6 ± 0.9 and 26.5 ± 0.1, respectively.
Fig. 4.
Effect of LH on FGF9-induced CCND1 (Panel A) and CDK4 (Panel B) mRNA expression in large-follicle TC (Exp. 4). Cells were cultured for 72 h as described in Materials and Methods, and then treated for an additional 24 h with IGF1 (30 ng/mL) and LH (0 or 30 ng/mL), FGF9 (0 or 30 ng/mL) or LH plus FGF9. Values are normalized to constitutively expressed 18S ribosomal RNA and are means ± SEM of three separate experiments. a,bMeans without a common letter differ (p < 0.05).
Experiment 5
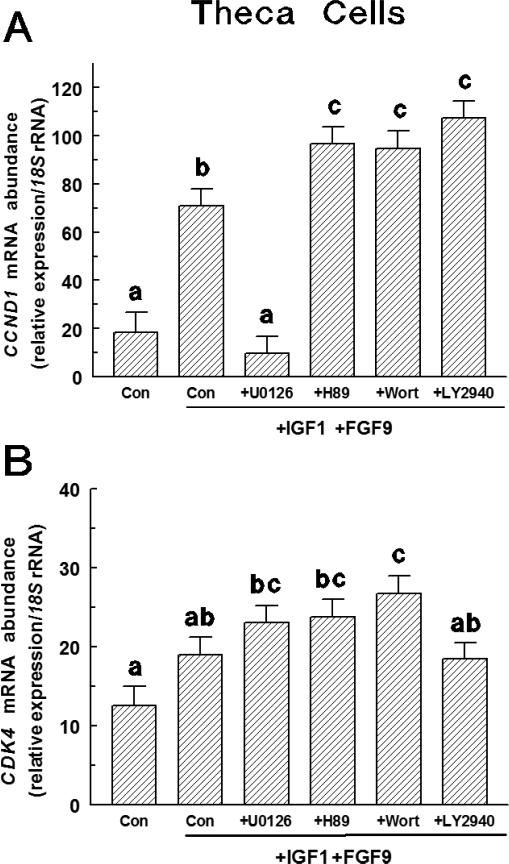
In large-follicle TC, FGF9 plus IGF1 treatment increased (p < 0.0001) CCND1 mRNA expression compared to controls (Figure 5A). U0126 (a MAPK/ERK inhibitor) reduced (p < 0.0001) CCND1 mRNA abundance compared to FGF9 plus IGF1-treated controls and did not differ from untreated controls. All other inhibitors increased (p < 0.05) CCND1 mRNA abundance compared to FGF9 plus IGF1-treated controls (Figure 5A). Abundance of CDK4 mRNA did not differ between untreated control and FGF9 plus IGF1-treated controls (Figure 5B), and U0126, LY294002 (a PI3K inhibitor) and H89 (a PKA inhibitor) had no effect (p < 0.10) on CDK4 mRNA abundance compared to FGF9 plus IGF1 controls (Figure 5B). Only wortmannin (a PI3K inhibitor) increased (p < 0.05) CDK4 mRNA abundance compared to FGF9 plus IGF1-treated controls. The CCND1 Ct and CDK4 Ct averaged 25.8 ± 0.1 and 29.1 ± 0.1, respectively.
Fig. 5.
Effect of various intracellular inhibitors on FGF9-induced CCND1 (Panel A) and CDK4 (Panel B) mRNA expression in large-follicle TC (Exp. 5). Cells were cultured for 72 h as described in Materials and Methods, and then treated for an additional 24 h with IGF1 (30 ng/mL) plus and FGF9 (30 ng/mL) and either LY294002, H89, U0126 or wortmannin. Values are normalized to constitutively expressed 18S ribosomal RNA and are means ± SEM of three separate experiments. a,b,cWithin a panel, means without a common letter differ (p < 0.05).
Experiment 6
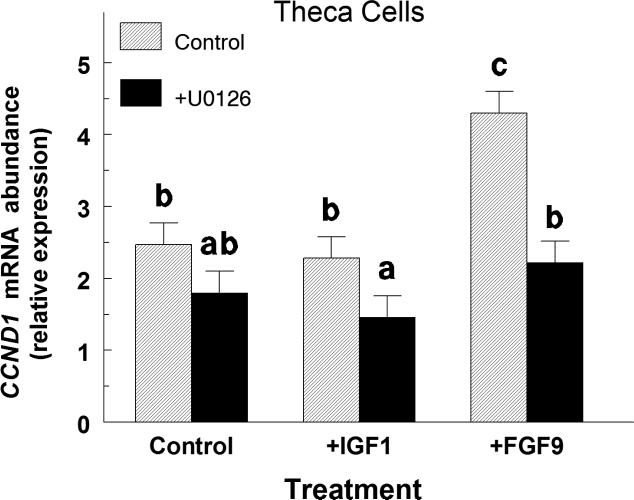
In large-follicle TC, IGF1 treatment alone did not affect (p > 0.10) CCND1 mRNA abundance whereas FGF9 increased (p < 0.05) CCND1 mRNA abundance by 1.7-fold (Figure 6). Addition of the MAPK inhibitor, U0126 completely blocked the FGF9-induced increase in CCND1 mRNA (Figure 5). The average CCND1 Ct was 26.4 ± 0.2.
Fig. 6.
Effect of U0126 on CCND1 mRNA expression in IGF1- and FGF9-treated large-follicle TC (Exp. 6). Cells were cultured for 96 h as described in Materials and Methods, and then treated for an additional 24 h with IGF1 (30 ng/ml) or FGF9 (30 ng/ml) with or without U0126 (20 μM). Values are normalized to constitutively expressed 18S ribosomal RNA and are means ± SEM of three separate experiments. a,b,cMeans without a common letter differ (p < 0.05).
Experiment 7
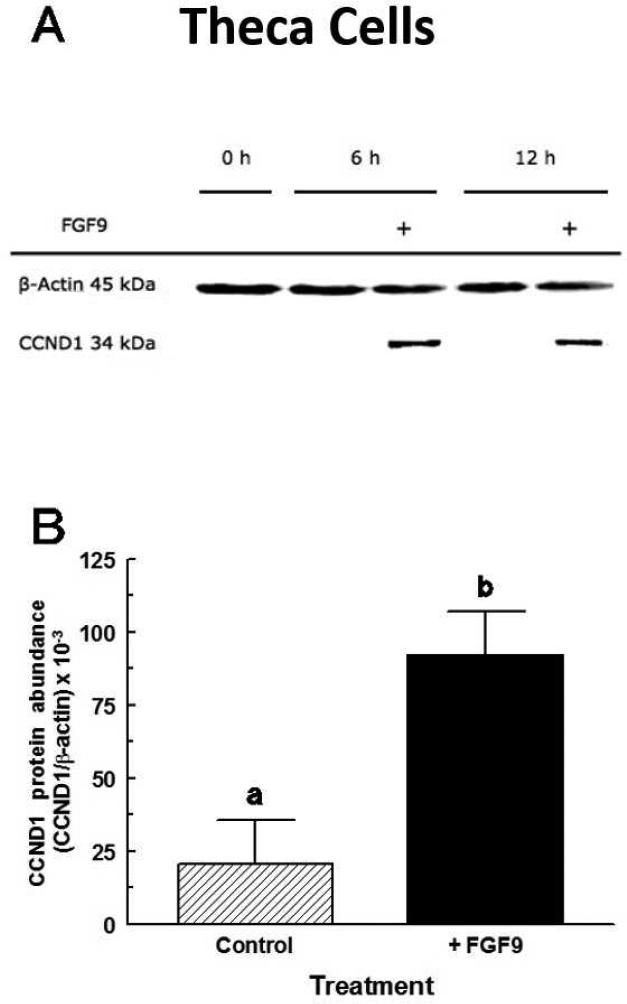
CCND1 protein levels in TC increased (p < 0.05) 4.5-fold between 0 h and 6 h after FGF9 treatment (Figure 6A). CCND1 protein levels at 6 h and 12 h (Figure 7A) did not differ (p > 0.10) so the 6 h and 12 h values were combined in Figure 7B. There was no significant effect of time or FGF9 treatment on CDK4 protein accumulation in TC (data not shown).
Fig. 7.
Stimulatory effect of FGF9 on CCND1 protein expression in large-follicle TC (Exp. 7). Cells were cultured for 96 h as described in Materials and Methods, and then treated for an additional 24 h in serum-free medium with either no addition (Control) or FGF9 (30 ng/mL). Panel A shows results from a representative Western blot. Panel B are means ± SEM of the 6 h and 12 h values averaged from three separate experiments. a,bMeans without a common letter differ (p < 0.05).
4. Discussion
FGF9 was first discovered in the rat ovary (Drummond et al., 2007) and later in the bovine ovary by Grado-Ahuir et al. (2011). Using microarray technology Grado-Ahuir et al., (2011) demonstrated a reduced expression of FGF9 mRNA in GC of cystic vs. normal dominant follicles. Since then, FGF9 has been shown to increase cell proliferation in both TC and GC of cattle (Schreiber and Spicer, 2012; Schreiber et al., 2012), yet how this increase is mediated was unknown. The results of the present study indicate that: (1) FGF9 treatment increased CCND1 mRNA and protein levels in TC; (2) FGF9 increased CCND1 mRNA expression in small and large follicle GC; (3) FGF9 did not alter CDK4 mRNA or protein levels in TC; and (4) FGF9 likely uses the MEK pathway to increase CCND1 mRNA expression. The precise point during the cell cycle that is stimulated by FGF9 will require further study but based on previous studies in other cell types suggest that stimulation of CCND1 could promote G1/S transition (Liu et al., 2014; Chang et al., 2015; Li et al., 2016).
Usually, CDK expression remains constant while cyclin expression fluctuates based on the point of the cell in the cycle, and preparedness to enter the next phase (Alberts et al., 2002). Consistent with this idea, we found that bovine TC treated with FGF9 had increased CCND1 mRNA abundance with no change in CDK4 mRNA expression. In addition, FGF9 increased CCND1 mRNA abundance in large- and small-follicle GC. Because CCND1 is a transiently and lowly expressed gene, TC were serum-starved for 24 h in order to get more cells in the G1 phase, and then stimulated with FGF9; after 6 and 12 h of FGF9 treatment CCND1 protein was significantly greater than control. Previously, CCND1 was thought to be of little importance in rats and mice because CCND1 mRNA was not expressed in GC and weakly expressed in TC (Robker and Richards, 1998). The only study characterizing CCND1 mRNA in bovine GC reported no change during an estrous cycle, while both CCND2 and CCND3 mRNA expression differed among phases of the estrous cycle (Shimizu et al., 2013). However, CCND1 mRNA is induced by GDF9 in human GC (Huang et al., 2009) and by EGF, FGF2 and ginsenosides in chicken GC (Tan et al., 2010; Lin et al., 2011; 2012). In endometrial tissue, FGF9 induces CCND1 (Schwartz et al., 2003; Wing et al., 2005; Chan et al., 2012). Thus, FGF9 induction of CCND1 in the present and previous studies would imply that the G1 interval of the cell cycle would be shortened, and thereby accelerate cell proliferation. Abundance of CDK4 mRNA was also increased by FGF9 treatment in large-follicle GC and tended to increase in small follicle GC. Although expression of CDKs usually remains constant throughout the cell cycle (Alberts et al., 2002), some treatments have been shown to alter their expression in vitro. Previously, CDK4 mRNA was altered by curcumin in rat hepatic cells (Huang et al., 2013) and by TGF-β in porcine valve interstitial cells (Li and Gotlieb, 2011). However, in bovine large-follicle TC, treatment of FGF9 did not significantly alter CDK4 mRNA expression or protein levels.
FGF9 has the ability to bind all FGFR, although some receptors bind FGF9 with higher affinity than others. Receptors that bind FGF9 with the greatest affinity are FGFR3c, −2c and −4 (in descending order); other FGFs that bind to these receptors are FGF1, −2, −4, −6, −8, −16, −17, −18, and −20 (Ornitz et al., 1996; Pownall and Isaacs, 2010). FGFs are able to exert their specific actions because different FGFRs are present and different FGF ligands are produced in each cell type and species. The FGFRs are transmembrane tyrosine kinase receptors that activate multiple signaling pathways including PLCγ, PI3K/AKT, and RAS/MAPK, with the latter being most prominent (Powers et al., 2000). Our inhibitor study revealed that the RAS/MAPK inhibitor, U0126, inhibited the FGF9+IGF1-induced increase of CCND1 mRNA expression, while all other inhibitors significantly up-regulated CCND1 mRNA expression over that of FGF9+IGF1 treatment. This suggests FGF9 uses the RAS/MAPK signaling pathway to induce CCND1 gene transcription and/or translation. In human endometrial stromal cells, PD98059 (a MEK inhibitor) abrogated FGF9-induced proliferation and CCND1 induction by FGF9 (Wing et al., 2005), and in mouse spermatogonia, U0126 abolished the FGF9-induced induction of pSmad2 and Cripto levels whereas LY294002 did not exert any effect (Tassinari et al., 2015). Wing et al., (2005) further concluded that PI3K is not involved in the FGF9-induced stromal cell proliferation and this is consistent with our studies in theca cells. In contrast, FGF9-induced testosterone by mouse Leydig cells was inhibited by wortmannin (a PI3K inhibitor), farnesyl protein transferase inhibitor III (a Ras inhibitor) and H89 (a PKA inhibitor) but not PLCγ inhibitors (Lin et al., 2010). Lai et al. (2014) further reported that FGF9 activates AKT and MAPK pathways to stimulate steroidogenesis in mouse Leydig cells. Thus, the precise downstream signal transduction mediator(s) of FGF9 may depend on the target gene(s) and cell type in which it is acting. The increase in CCND1 mRNA expression after treatment with H89, wortmannin and LY294002 may be the result of more intracellular signaling via the RAS/MAPK pathway when other pathways are inhibited. Additionally, the increase in CCND1 mRNA expression in response to FGF9+IGF1 treatment appears to be attributed to FGF9 as well as its interaction with IGF1, since CCND1 response to FGF9 was twofold greater in the presence than in the absence of IGF1. Because IGF1 alone had no effect on CCND1 mRNA abundance in the present study and IGF1 uses the PI3K/AKT pathway (Wing et al., 2005; Mani et al., 2010), the synergism between FGF9 and IGF1 is likely do to amplification of the FGF9 intracellular signaling pathway. Furthermore, when the IGF1 pathway was inhibited by wortmannin or LY294002, FGF9+IGF1-induced CCND1 mRNA expression did not decrease.
5. Conclusion
Taken together these results indicate the FGF9-induced increase in GC and TC proliferation is mediated by rapid up-regulation of CCND1 gene expression and protein translation. The intracellular pathway of FGF9-induced CCND1 appears to be via RAS/MAPK. There is still much unknown about FGF9's involvement in follicular function and cell cycle progression, these findings will help us better understand the intracellular targets of FGF9 in bovine ovarian cells.
Highlights.
FGF9 increases CCND1 mRNA and protein in bovine theca cells from large follicles.
FGF9 increases CCND1 mRNA in granulosa cell from small and large follicles.
IGF1 amplifies the FGF9-induced CCND1 mRNA in granulosa cells from small follicles.
FGF9 likely uses the MEK pathway to increase CCND1 mRNA expression.
Acknowledgments
The authors thank: Dr. Steve Hartson, Department of Biochemistry and Molecular Biology for helpful discussions regarding CCND1 Western blotting; Dr. J. Hernandez-Gifford and B. Parker for assistance with CDK4 and β-actin Western blotting; N. Schreiber, J. Evans, J. Williams, P. Aad, L. F. Schutz, L. Zhang, and A. Stapp for technical assistance; the OSU Microarray Core Facility and OSU Recombinant DNA/Protein Resource Facility for use of equipment; Dr. A. F. Parlow, National Hormone & Pituitary Program, (Torrance, CA) for purified LH and FSH; and Creekstone Farms (Arkansas City, KS) for their generous donations of bovine ovaries. Approved for publication by the Director, Oklahoma Agric. Exp. Sta., and supported in part by: the NICHD, National Institutes of Health, through Agreement R15-HD-066302, and the Oklahoma State University Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
All of the authors confirm that there is no conflict of interest in relation to this work.
References
- Aad PY, Voge JL, Santiago CA, Malayer JR, Spicer LJ. Real-time RT-PCR quantification of pregnancy-associated plasma protein-A mRNA abundance in bovine granulosa and theca cells: effects of hormones in vitro. Domest. Anim. Endocrinol. 2006;31:357–372. doi: 10.1016/j.domaniend.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell. 4th ed. Garland Science; New York, NY.: 2002. [Google Scholar]
- Asselin E, Wang Y, Tsang BK. X-linked inhibitor of apoptosis protein activates the phosphatidylinositol 3-kinase/Akt pathway in rat granulosa cells during follicular development. Endocrinology. 2001;142:2451–2457. doi: 10.1210/endo.142.6.8080. [DOI] [PubMed] [Google Scholar]
- Bendris N, Lemmers B, Blanchard JM. Cell cycle, cytoskeleton dynamics and beyond: the many functions of cyclins and CDK inhibitors. Cell Cycle. 2015;14:1786–1798. doi: 10.1080/15384101.2014.998085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisha B, Schams D, Kosmann M, Amselgruber W, Einspanier R. Expression and localisation of vascular endothelial growth factor and basic fibroblast growth factor during the final growth of bovine ovarian follicles. J. Endocrinol. 2000;167:371–382. doi: 10.1677/joe.0.1670371. [DOI] [PubMed] [Google Scholar]
- Berisha B, Sinowatz F, Schams D. Expression and localization of fibroblast growth factor (FGF) family members during the final growth of bovine ovarian follicles. Mol. Reprod. Dev. 2004;67:162–171. doi: 10.1002/mrd.10386. [DOI] [PubMed] [Google Scholar]
- Buratini J, Jr., Pinto MG, Castilho AC, Amorim RL, Giometti IC, Portela VM, Nicola ES, Price CA. Expression and function of fibroblast growth factor 10 and its receptor, fibroblast growth factor receptor 2B, in bovine follicles. Biol. Reprod. 2007;77:743–750. doi: 10.1095/biolreprod.107.062273. [DOI] [PubMed] [Google Scholar]
- Buratini J, Jr., Teixeira AB, Costa IB, Glapinski VF, Pinto MG, Giometti IC, Barros CM, Cao M, Nicola ES, Price CA. Expression of fibroblast growth factor-8 and regulation of cognate receptors, fibroblast growth factor receptor-3c and -4, in bovine antral follicles. Reproduction. 2005;130:343–350. doi: 10.1530/rep.1.00642. [DOI] [PubMed] [Google Scholar]
- Castañon BI, Stapp AD, Gifford CA, Spicer LJ, Hallford DM, Hernandez Gifford JA. Follicle-stimulating hormone regulation of estradiol production: possible involvement of WNT2 and β-catenin in bovine granulosa cells. J. Anim. Sci. 2012;90:3789–3797. doi: 10.2527/jas.2011-4696. [DOI] [PubMed] [Google Scholar]
- Chan DW, Mak CS, Leung TH, Chan KK, Ngan HY. Down-regulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget. 2012;3:1546–56. doi: 10.18632/oncotarget.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Niu Z, Gu N, Zhao W, Wang G, Jia Y, Li D, Xu C. Analysis of the ways and methods of signaling pathways in regulating cell cycle of NIH3T3 at transcriptional level. BMC Cell Biol. 2015;16:25. doi: 10.1186/s12860-015-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves RN, de Matos MH, Buratini J, Jr., de Figueiredo JR. The fibroblast growth factor family: involvement in the regulation of folliculogenesis. Reprod. Fertil. Dev. 2012;24:905–915. doi: 10.1071/RD11318. [DOI] [PubMed] [Google Scholar]
- Dewi DA, Abayasekara DR, Wheeler-Jones CP. Requirement for ERK1/2 activation in the regulation of progesterone production in human granulosa-lutein cells is stimulus specific. Endocrinology. 2002;143:877–888. doi: 10.1210/endo.143.3.8677. [DOI] [PubMed] [Google Scholar]
- Drummond AE, Tellbach M, Dyson M, Findlay JK. Fibroblast growth factor-9, a local regulator of ovarian function. Endocrinology. 2007;148:3711–3721. doi: 10.1210/en.2006-1668. [DOI] [PubMed] [Google Scholar]
- Grado-Ahuir JA, Aad PY, Spicer LJ. New insights into the pathogenesis of cystic follicles in cattle: microarray analysis of gene expression in granulosa cells. J. Anim. Sci. 2011;89:1769–1786. doi: 10.2527/jas.2010-3463. [DOI] [PubMed] [Google Scholar]
- Huang CZ, Huang WZ, Zhang G, Tang DL. In vivo study on the effects of curcumin on the expression profiles of anti-tumour genes (VEGF, CyclinD1 and CDK4) in liver of rats injected with DEN. Mol. Biol. Rep. 2013;40:5825–5831. doi: 10.1007/s11033-013-2688-y. [DOI] [PubMed] [Google Scholar]
- Huang Q, Cheung AP, Zhang Y, Huang HF, Auersperg N, Leung PC. Effects of growth differentiation factor 9 on cell cycle regulators and ERK42/44 in human granulosa cell proliferation. Am. J. Physiol. Endocrinol. Metab. 2009;296:E1344–E1353. doi: 10.1152/ajpendo.90929.2008. [DOI] [PubMed] [Google Scholar]
- Laestander C, Engström W. Role of fibroblast growth factors in elicitation of cell responses. Cell. Prolif. 2014;47:3–11. doi: 10.1111/cpr.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagaly DV, Aad PY, Grado-Ahuir JA, Hulsey LB, Spicer LJ. Role of adiponectin in regulating ovarian theca and granulosa cell function. Mol. Cell. Endocrinol. 2008;284:38–45. doi: 10.1016/j.mce.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Lai MS, Cheng YS, Chen PR, Tsai SJ, Huang BM. Fibroblast growth factor 9 activates akt and MAPK pathways to stimulate steroidogenesis in mouse Leydig cells. PLoS One. 2014;9:e90243. doi: 10.1371/journal.pone.0090243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhout DJ, Spicer LJ, Geisert RD. Development of a culture system for bovine GC: effects of growth hormone, estradiol, and gonadotrophins on cell proliferation, steroidogenesis, and protein synthesis. J. Anim. Sci. 1991;69:3321–3334. doi: 10.2527/1991.6983321x. [DOI] [PubMed] [Google Scholar]
- Laurich VM, Trbovich AM, O'Neill FH, Houk CP, Sluss PM, Payne AH, Donahoe PK, Teixeira J. Müllerian inhibiting substance blocks the protein kinase A-induced expression of cytochrome p450 17alpha-hydroxylase/C(17-20) lyase mRNA in a mouse Leydig cell line independent of cAMP responsive element binding protein phosphorylation. Endocrinology. 2002;143:3351–3360. doi: 10.1210/en.2001-211352. [DOI] [PubMed] [Google Scholar]
- Li C, Gotlieb AI. Transforming growth factor-β regulates the growth of valve interstitial cells in vitro. Am. J. Pathol. 2011;179:1746–1755. doi: 10.1016/j.ajpath.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li D, Yang W, Fu H, Liu Y, Li Y. Overexpression of the transcription factor FOXP3 in lung adenocarcinoma sustains malignant character by promoting G1/S transition gene CCND1. Tumour Biol. 2016;37:7395–404. doi: 10.1007/s13277-015-4616-3. [DOI] [PubMed] [Google Scholar]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Lin J, Jia Y, Zhang C. Effect of epidermal growth factor on follicle-stimulating hormone-induced proliferation of granulosa cells from chicken prehierarchical follicles. J. Zhejiang Univ. Sci. B. 2011;12:875–883. doi: 10.1631/jzus.B1100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Jia Y, Zeng W, Mi Y, Zhang C. Basic FGF promotes proliferation of ovarian granulosa cells in the laying chickens via FGFR1 and PKC pathway. Reprod. Domest. Anim. 2012;47:135–142. doi: 10.1111/j.1439-0531.2011.01813.x. [DOI] [PubMed] [Google Scholar]
- Lin YM, Tsai CC, Chung CL, Chen PR, Sun HS, Tsai SJ, Huang BM. Fibroblast growth factor 9 stimulates steroidogenesis in postnatal Leydig cells. Int. J. Androl. 2010;33:545–553. doi: 10.1111/j.1365-2605.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang H, Shi L, Zhang W, Yuan J, Chen X, Liu J, Zhang Y, Wang Z. Inhibition of Rac1 activity induces G1/S phase arrest through the GSK3/cyclin D1 pathway in human cancer cells. Oncol. Rep. 2014;32:1395–400. doi: 10.3892/or.2014.3388. [DOI] [PubMed] [Google Scholar]
- Machado MF, Portela VM, Price CA, Costa IB, Ripamonte P, Amorim RL, Buratini J., Jr. Regulation and action of fibroblast growth factor 17 in bovine follicles. J. Endocrinol. 2009;202:347–53. doi: 10.1677/JOE-09-0145. [DOI] [PubMed] [Google Scholar]
- Mani AM, Fenwick MA, Cheng Z, Sharma MK, Singh D, Wathes DC. IGF1 induces up-regulation of steroidogenic and apoptotic regulatory genes via activation of phosphatidylinositol-dependent kinase/AKT in bovine granulosa cells. Reproduction. 2010;139:139–151. doi: 10.1530/REP-09-0050. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Ott L. An Introduction to Statistical Methods and Data Analysis. Duxbury Press; North Scituate, MA.: 1977. p. 384. [Google Scholar]
- Parrott JA, Skinner MK. Developmental and hormonal regulation of keratinocyte growth factor expression and action in the ovarian follicle. Endocrinology. 1998;139:228–235. doi: 10.1210/endo.139.1.5680. [DOI] [PubMed] [Google Scholar]
- Poretsky L, Seto-Young D, Shrestha A, Dhillon S, Mirjany M, Liu HC, Yih MC, Rosenwaks Z. Phosphatidyl-inositol-3 kinase-independent insulin action pathway(s) in the human ovary. J. Clin. Endocrinol. Metab. 2001;86:3115–3119. doi: 10.1210/jcem.86.7.7617. [DOI] [PubMed] [Google Scholar]
- Portela VM, Machado M, Buratini J, Jr., Zamberlam G, Amorim RL, Goncalves P, Price CA. Expression and function of fibroblast growth factor 18 in the ovarian follicle in cattle. Biol. Reprod. 2010;83:339–346. doi: 10.1095/biolreprod.110.084277. [DOI] [PubMed] [Google Scholar]
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors, and signaling. Endocr. Relat. Cancer. 2000;7:165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Isaacs HV. Morgan and Claypool Life Sciences. San Rafael, CA.: 2010. FGF signaling in Vertebrate Development. [PubMed] [Google Scholar]
- Robker RL, Richards JS. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol. Endocrinol. 1998;12:924–940. doi: 10.1210/mend.12.7.0138. [DOI] [PubMed] [Google Scholar]
- Schreiber NB, Spicer LJ. Effects of fibroblast growth factor 9 (FGF9) on steroidogenesis and gene expression and control of FGF9 mRNA in bovine granulosa cells. Endocrinology. 2012;153:4491–4501. doi: 10.1210/en.2012-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber NB, Totty ML, Spicer LJ. Expression and effect of fibroblast growth factor 9 in bovine theca cells. J. Endocrinol. 2012;215:167–175. doi: 10.1530/JOE-12-0293. [DOI] [PubMed] [Google Scholar]
- Schwartz DR, Wu R, Kardia SL, Levin AM, Huang CC, Shedden KA, Kuick R, Misek DE, Hanash SM, Taylor JM, Reed H, Hendrix N, Zhai Y, Fearon ER, Cho KR. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- Shimizu T, Hirai Y, Miyamoto A. Expression of cyclins and cyclin-dependent kinase inhibitors in granulosa cells from bovine ovary. Reprod. Domest. Anim. 2013;48:e65–e69. doi: 10.1111/rda.12177. [DOI] [PubMed] [Google Scholar]
- Silva JM, Hamel M, Sahmi M, Price CA. Control of oestradiol secretion and of cytochrome P450 aromatase messenger ribonucleic acid accumulation by FSH involves different intracellular pathways in oestrogenic bovine granulosa cells in vitro. Reproduction. 2006;132:909–917. doi: 10.1530/REP-06-0058. [DOI] [PubMed] [Google Scholar]
- Spicer LJ, Aad PY. Insulin-like growth factor (IGF) 2 stimulates steroidogenesis and mitosis of bovine granulosa cells through the IGF1 receptor: role of follicle-stimulating hormone and IGF2 receptor. Biol. Reprod. 2007;77:18–27. doi: 10.1095/biolreprod.106.058230. [DOI] [PubMed] [Google Scholar]
- Spicer LJ, Chamberlain CS, Maciel SM. Influence of gonadotropins on insulin- and insulin-like growth factor-I (IGF1)-induced steroid production by bovine granulosa cells. Domest. Anim. Endocrinol. 2002;22:237–254. doi: 10.1016/s0739-7240(02)00125-x. [DOI] [PubMed] [Google Scholar]
- Spicer LJ, Schreiber NB, Lagaly DV, Aad PY, Douthit LB, Grado-Ahuir JA. Effect of resistin on granulosa and theca cell function in cattle. Anim. Reprod. Sci. 2001;124:19–27. doi: 10.1016/j.anireprosci.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Spicer LJ, Sudo S, Aad PY, Wang LS, Chun SY, Ben-Shlomo I, Klein C, Hsueh AJ. The hedgehog-patched signaling pathway and function in the mammalian ovary: a novel role for hedgehog proteins in stimulating proliferation and steroidogenesis of theca cells. Reproduction. 2009;138:329–339. doi: 10.1530/REP-08-0317. [DOI] [PubMed] [Google Scholar]
- Stapp AD, Gómez BI, Gifford CA, Hallford DM, Hernandez Gifford JA. Canonical WNT signaling inhibits follicle stimulating hormone mediated steroidogenesis in primary cultures of rat granulosa cells. PLoS One. 2014;9:e86432. doi: 10.1371/journal.pone.0086432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TQ, Ge C, Mi Y, Zhang C. Ginsenosides promote proliferation of granulosa cells from chicken prehierarchical follicles through PKC activation and up-regulated cyclin gene expression. Cell Biol. Int. 2010;34:769–775. doi: 10.1042/CBI20090244. [DOI] [PubMed] [Google Scholar]
- Tassinari V, Campolo F, Cesarini V, Todaro F, Dolci S, Rossi P. Fgf9 inhibition of meiotic differentiation in spermatogonia is mediated by Erk-dependent activation of Nodal-Smad2/3 signaling and is antagonized by Kit Ligand. Cell Death Dis. 2015;6:e1688. doi: 10.1038/cddis.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel IL, Umapathysivam K, Tilley WD, Rodgers RJ. Immunohistochemical localization of basic fibroblast growth factor in bovine ovarian follicles. Mol. Cell. Endocrinol. 1995;115:133–140. doi: 10.1016/0303-7207(95)03678-4. [DOI] [PubMed] [Google Scholar]
- Voge JL, Aad PY, Santiago CA, Goad DW, Malayer JR, Allen D, Spicer LJ. Effect of insulin-like growth factors (IGF), FSH, and leptin on IGF-binding-protein mRNA expression in bovine granulosa and theca cells: quantitative detection by real-time PCR. Peptides. 2004;25:2195–2203. doi: 10.1016/j.peptides.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Wing LY, Chen HM, Chuang PC, Wu MH, Tsai SJ. The mammalian target of rapamycin-p70 ribosomal S6 kinase but not phosphatidylinositol 3-kinase-Akt signaling is responsible for fibroblast growth factor-9-induced cell proliferation. J. Biol. Chem. 2005;280:19937–19947. doi: 10.1074/jbc.M411865200. [DOI] [PubMed] [Google Scholar]