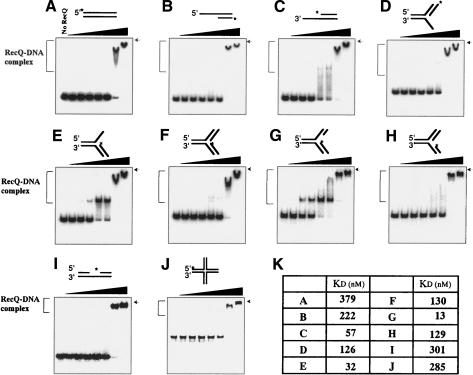
Figure 1.
Gel mobility shift analysis of RecQ binding to forked DNA structures. Blunt-end (A), 5′-overhang (B), 3′-overhang (C), 5′-flap (D), 3′-flap (E), forked DNA with no gap (F), LeGF DNA (G), LaGF DNA (H), gapped DNA (I), or HJ DNA (J; 10 nM) was used in binding reactions. RecQ was added at various concentrations (1 nM, 5 nM, 10 nM, 50 nM, 100 nM, 500 nM, and 1000 nM) in the absence of ATP. Protein–DNA complexes were analyzed by 6% nondenaturing PAGE. (*) 32P-labeling. The position of the well on the gel is indicated by arrows. DNA-binding reactions were performed as described in Materials and Methods. (K) Summary of dissociation constants of RecQ with various forms of DNA substrates.