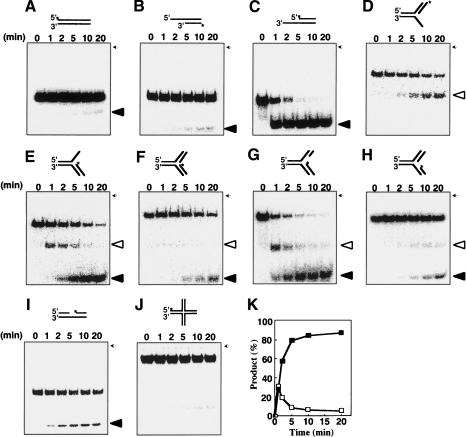
Figure 3.
Time course analysis of RecQ-catalyzed DNA unwinding activity. (A–J) RecQ was incubated at 26°C with blunt-ended (A), 5′-overhang (B), 3′-overhang (C), 5′-flap (D), 3′-flap (E), forked DNA with no gap (F), LeGF DNA (G), LaGF DNA (H), duplex DNA with a gap (I), or HJ DNA (J). Reactions containing 10 nM RecQ, 10 nM DNA, and 1 mM ATP were carried out at 26°C and initiated by the addition of ATP. Aliquots were taken at 1, 2, 5, 10, and 20 min. Deproteinized reaction products except the HJ DNA (9%) were analyzed by 12% nondenaturing PAGE. Open triangles indicate 3′- or 5′-overhang DNA, and closed triangles indicate 32P-labeled ssDNA. (K) The reaction products from G were quantified using a PhosphorImager (Fuji BAS1500) and plotted. Open and filled symbols represent 3′-overhang and 30-mer nascent lagging-strand DNA, respectively. (*) 32P-labeling. The position of the well on the gel is indicated by arrows.