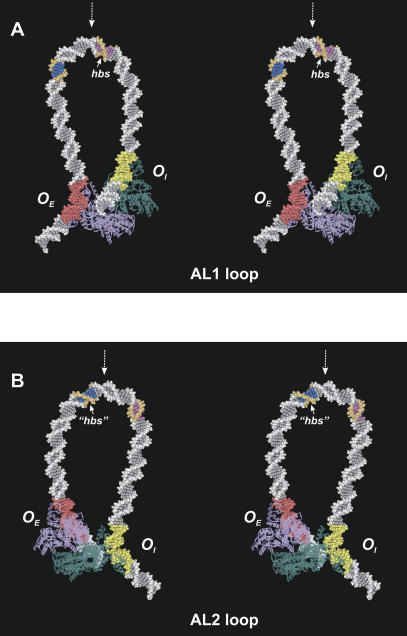
Figure 6.
Stereoviews of the minimal energy configurations of the loop modes AL1 (A, back view) and AL2 (B, front view). Two GalR dimers (purple and teal blue) are shown as ribbons. The OE operator is highlighted with red color, and the OI is highlighted with yellow. The -10 element of the P2 promoter is colored blue and orange. The AL1 and AL2 loops have similar overall structure. However, the direction of local DNA bending is different in two modes; the DNA surface that is inside at the AL1 loop apex is turned halfway outside in the AL2 loop (cf. orientation of the DNA grooves in the colored DNA fragments). The experimentally observed HU-binding site (hbs, position +6.5, 6 bp are colored magenta and orange) and putative binding site (“hbs”, position -14.5) occupy structurally equivalent segments of the AL1 and AL2 loops relative to the corresponding apex (indicated with dashed white arrows).