Abstract
Resistance to 5-flucytosine (5-FC), used as an antifungal drug in combination therapy, compromises its therapeutic action. In this work, the response of the human pathogen Candida glabrata to 5-FC was evaluated at the membrane proteome level, using an iTRAQ-based approach. A total of 32 proteins were found to display significant expression changes in the membrane fraction of cells upon exposure to 5-FC, 50% of which under the control of CgPdr1, the major regulator of azole drug resistance. These proteins cluster into functional groups associated to cell wall assembly, lipid metabolism, amino acid/nucleotide metabolism, ribosome components and translation machinery, mitochondrial function, glucose metabolism, and multidrug resistance transport. Given the obtained indications, the function of the drug:H+ antiporters CgFlr1 (ORF CAGL0H06017g) and CgFlr2 (ORF CAGL0H06039g) was evaluated. The expression of both proteins, localized to the plasma membrane, was found to confer flucytosine resistance. CgFlr2 further confers azole drug resistance. The deletion of CgFLR1 or CgFLR2 was seen to increase the intracellular accumulation of 5-FC, or 5-FC and clotrimazole, suggesting that these transporters play direct roles in drug extrusion. The expression of CgFLR1 and CgFLR2 was found to be controlled by the transcription factors CgPdr1 and CgYap1, major regulator of oxidative stress resistance.
Keywords: antifungal drug resistance, flucytosine, CgFlr1 and CgFlr2, Candida glabrata, CgPdr1
Introduction
Systemic fungal infections are a problem of increasing clinical significance, especially since the prophylactic and therapeutic use of antifungal drugs has led to an increased number of infections with drug resistant fungal pathogens (Fidel et al., 1999; Mishra et al., 2007).
The antifungal drug 5-flucytosine (5-FC) is a fluorinated pyrimidine which enters fungal cells through permeases (Ghannoum and Rice, 1999; Hope et al., 2004; Edlind and Katiyar, 2010) and is then converted, by cytosine deaminase, to its metabolically active form 5-fluorouracil (5-FU) (Ghannoum and Rice, 1999; Espinel-Ingroff, 2008; Edlind and Katiyar, 2010). This antifungal drug acts by inhibiting transcription, DNA replication and protein synthesis (Ghannoum and Rice, 1999; Espinel-Ingroff, 2008). The specificity of this antimycotic relies on the absence of cytosine deaminase in mammalian cells (Hope et al., 2004; Edlind and Katiyar, 2010). However, 5-FU is considered toxic, mostly due to the conversion of flucytosine to fluorouracil by gut bacteria (Vermes et al., 2000). Despite these side-effects, 5-FC is still used clinically, mostly in combination therapy (Espinel-Ingroff, 2008).
Resistance to 5-FC in clinically relevant Candida species develops rapidly under treatment (Sanglard and Odds, 2002). Resistance is often related to decreased drug uptake by the Fcy2 cytosine permease or decreased conversion of 5-FC to 5-FU or 5-FUMP by the Fcy1 and Fur1 enzymes (Kontoyiannis and Lewis, 2002; Papon et al., 2007; Espinel-Ingroff, 2008). Some epidemiological studies suggest, however, that resistance mechanisms, independent of the Fcy2-Fcy1-Fur1 pathway, may play an important role in this phenomenon (Zhang et al., 2002). For example, reduction of 5-FC intracellular accumulation, mediated by the Drug:H+ Antiporter (DHA) CgAqr1 (Costa et al., 2013a), or by the acquaglyceroporins CgFps1 and CgFps2 (Costa et al., 2015) were recently registered.
Given these observations, in this study the Candida glabrata response to 5-FC was analyzed at the membrane proteome level, using an iTRAQ-based approach. Among the obtained results, the concentration of the DHA transporter CgFlr1 was seen to increase in 5-FC challenged cells. The role of CgFlr1 (ORF CAGL0H06017g), and of its very close homolog CgFlr2 (ORF CAGL0H06039g) (Gbelska et al., 2006; Costa et al., 2014a), in the resistance to 5-FC was then analyzed.
The deletion of CgFLR1 had been found to lead to increased susceptibility to benomyl, diamide, and menadione, but not to fluconazole (Chen et al., 2007). As for CgFlr2, it remained uncharacterized until this study (Costa et al., 2014a). Both proteins, however, are close homologs of the Saccharomyces cerevisiae Flr1 DHA transporter. Flr1 confers resistance to many unrelated chemical compounds as reviewed in Sá-Correia et al. (2009), its expression being highly responsive to chemical stress exposure (Brôco et al., 1999; Teixeira et al., 2008, 2010; Sá-Correia et al., 2009; Dos Santos et al., 2014). The FLR1 homolog in Candida albicans, MDR1, has been one of the few DHA transporters linked so far to azole drug resistance (White, 1997; Alarco and Raymond, 1999; Costa et al., 2013a,b, 2014a,b), being an important determinant of clinical acquisition of resistance against these antifungals (White, 1997; Alarco and Raymond, 1999; Costa et al., 2014a). These previous findings were used to guide the functional analysis of CgFlr1 and CgFlr2 in this study, in which the sub-cellular localization, role in antifungal drug resistance, and expression analysis was carried out.
Materials and methods
Strains and growth media
C. glabrata parental strain KUE100 (Ueno et al., 2007) and derived single deletion mutants KUE100_Δcgflr1 or KUE100_Δcgflr2, constructed in this study, as well as the C. glabrata strains 66032u and 66032u_Δcgpdr1 (Vermitsky and Edlind, 2004), provided by Thomas Edlind, from Drexel University, College of Medicine, Philadelphia, PA, were batch-cultured at 30°C, with orbital agitation (250 rpm) in basal medium (BM), with the following composition (per liter): 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco), 20 g glucose (Merck) and 2.65 g (NH4)2SO4 (Merck). C. glabrata strains L5U1 (cgura3Δ0, cgleu2Δ0), 84u (cgura3Δ0), and 84u_Δcgyap1, kindly provided by John Bennett, (Chen et al., 2007) from the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, USA, were grown in BM medium supplemented with 20 mg/L uracil and 60mg/L leucine. S. cerevisiae parental strain BY4741 (MATa, ura3Δ0, leu2Δ0, his3Δ1, met15Δ0) and the derived single deletion mutant BY4741_Δflr1 were obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). Cells were batch-cultured at 30°C, with orbital agitation (250 rpm) in supplemented with 20 mg/L methionine, 20 mg/L histidine, 60 mg/L leucine, 20 mg/L uracil (all from Sigma). Solid media contained, besides the above-indicated ingredients, 20 g/L agar (Iberagar). The plasmid pGREG576 was obtained from the Drag&Drop collection (Jansen et al., 2005).
Membrane proteome-wide analysis of C. glabrata response to 5-flucytosine
Exponentially growing wild-type 66032 C. glabrata strain and the derived 66032_Δcgpdr1 deletion mutant were transferred to fresh BM medium in the absence of stress (control conditions) or in the presence of 4 μg/mL 5-flucytosine (Sigma). Upon 1 h of cultivation, cells were harvested by centrifugation and the membrane protein fraction was obtained as described before (Pais et al., 2016). Expression proteomics analysis of the obtained membrane-enriched fraction was carried out using an iTRAQ-MS procedure, carried out as a paid service at the Keck Foundation Biotechnology Resource Laboratory, Yale University, USA (http://medicine.yale.edu/keck/proteomics/index.aspx), using the method followed in Pais et al. (2016). Briefly, samples were sonicated and proteins reduced by adding 50 mM TCEP (tris(2-carboxyethyl)phosphine), followed by 200 mM MMTS (methyl methane thiosulfonate). Protein digestion was achieved by adding 10 μL of a solution of 1 mg/mL Lys-C, followed by incubation at 37°C for 3 h, and 10 μL of 1 mg/mL trypsin, followed by overnight incubation at 37°C. Macro-spin desalt of the digests with C18 spin columns for cleanup and quantitation was carried out, followed by dissolution in 65 μL of 500 mM TEAB. iTRAQ labeling was carried out based on the AAA quant protocol. iTRAQ experiments were carried out through the SCX cartridge and experiments run on 5600.
The search parameters and acceptance criteria used were the following: Peaklist generating software: ProteinPilot 4.5 and Mascot; Search engine: Paragon Search Engine (ProteinPilot 4.2); Sequence Database/spectral library: C. glabrata [5478] from SwissProt (May 2013); The database used was downloaded from UniProt, with a total of 5197 protein entries. Mass spectrometric analysis is done on an AB SCIEX TripleTOF® 5600 mass spectrometer with AB SCIEX ProteinPilot™ software used for protein identification and quantitation. ProteinPilot utilizes a Paragon™ algorithm with hybrid sequence tag and feature probability database searches. Hence, specific details such as mass tolerances, specific modifications etc., are not utilized. All iTRAQ results are uploaded into the Yale Protein Expression database (YPED) for investigator viewing. Protein identification was considered reliable for a Protein Score >2, corresponding to a confidence level of 99%. A reserve decoy database search, followed by filtering of all peptides above 1% False Discovery Rate was carried out before protein grouping.
Proteomics data analysis started from 3 iTRAQ sets. The samples present in each of the sets were randomized to prevent bias, and in different sets distinct labels were used to tag the samples, ensuring that protein identification in the MS step is not biased by the tags. For each sample in a given set, protein quantification was only considered for p < 0.05. Protein expression changes above 1.5-fold or below 0.66-fold were considered relevant. Protein classification into functional groups was achieved based on their predicted function, according to the Candida Genome Database (www.candidagenome.org), or based on the function of their closest S. cerevisiae homolog, according to the Saccharomyces Genome Database (www.yeastgenome.org).
Disruption of the CgFLR1 and CgFLR2 genes
The deletion of the C. glabrata FLR1 and FLR2 genes (ORFs CAGL0H06017g and CAGL0H06039g, respectively) was carried out in the parental strain KUE100, using the method described by Ueno et al. (2011). The target genes CgFLR1 and CgFLR2 were replaced by a DNA cassette including the CgHIS3 gene, through homologous recombination. The replacement cassette was prepared by PCR using the primers 5′ AGAGAAAAATAAAACCAATTCTAAAACCAAATCCATATTACAACCCAATTGCAAAGGGCCGCTGATCACG-3′ and 5′-AATGTTAGTGTGAACTTGAATGTTAGATTTTCACGTGAATGAGAACTGAGAAATCACATCGTGAGGCTGG-3′, for the CgFLR1 gene, and the primers 5′-AGAATCATATTCATAAAGGTAACAAAACTACACAACAAATTATTAACTATTTTACAGGCCGCTGATCACG-3′ and 5′-AAATAATTTGTTCGGGGTAAGCACAATTGGAGGCTCTATCTTTTTTCTCTTCTTCACATCGTGAGGCTGG-3′, for the CgFLR2 gene. The pHIS906 plasmid including CgHIS3 was used as a template and transformation was performed as described previously (Ueno et al., 2007). Recombination locus and gene deletion were verified by PCR using the following pairs of primers: 5′-GAGGTGCTTAATATCGTCAC -3′ and 5′-CAACAACGTGTCCTACATG-3′; and 5′-GTGCATTTCAGGACACACT-3′ and 5′-GTATTTGTTCTTGTCCTGGTGTG -3′, respectively.
Cloning of the C. glabrata CgFLR1 and CgFLR2 genes (ORFs CAGL0H06017g and CAGL0H06039g, respectively)
The pGREG576 plasmid from the Drag & Drop collection was used to clone and express the C. glabrata ORFs CAGL0H06017g and CAGL0H06039g in S. cerevisiae, as described before for other heterologous genes (Costa et al., 2013a,b, 2014b). CgFLR1 or CgFLR2 DNA was generated by PCR, using genomic DNA extracted from the sequenced CBS138 C. glabrata strain, and the following specific primers: 5′–GAATTCGATATCAAGCTTATCGATACCGTCGACAGCAAAGATGAATTATCTTC -3′ and 5′-GCGTGACATAACTAATTACATGACTCGAGGTCGACTCACCTGTTGTATTTAGACATGG -3′; or 5′-GAATTCGATATCAAGCTTATCGATACCGTCGACAATGTATATCGGTGCATTTCAGGAC -3′ and 5′-GCGTGACATAACTAATTACATGACTCGAGGTCGACTCATGAATCTGGACTAAATCTTG -3′, respectively. These plasmids include a S. cerevisiae CEN/ARS element which had been shown before to work in C. glabrata (Willins et al., 2002). The GAL1 promoter present in the pGREG576_CgFLR1 and pGREG576_CgFLR2 plasmids was then replaced by the copper-induced MTI C. glabrata promoter, giving rise to the pGREG576_MTI_CgFLR1 and pGREG576_MTI_CgFLR2 plasmids. The MTI promoter DNA was generated by PCR, using genomic DNA extracted from the sequenced CBS138 C. glabrata strain, and the following specific primers: 5′-TTAACCCTCACTAAAGGGAACAAAAGCTGGAGCTCTGTACGACACGCATCATGTGGCAATC -3′ and 5′-GAAAAGTTCTTCTCCTTTACTCATACTAGTGCGGCTGTGTTTGTTTTTGTATGTGTTTGTTG -3′. The recombinant plasmids pGREG576_CgFLR1, pGREG576_CgFLR2, pGREG576_MTI_CgFLR1, and pGREG576_MTI_CgFLR2 were obtained through homologous recombination in S. cerevisiae and verified by DNA sequencing. As before (Costa et al., 2013a,b, 2014b), the transformation of L5U1 C. glabrata cells with the pGREG576 plasmids, as well as plasmid propagation was ensured by growth in selective uracil depleted medium.
CgFlr1 and CgFlr2 subcellular localization assessment
The sub-cellular localization of the CgFlr1 and CgFlr2 proteins was determined based on the observation of BY4741 S. cerevisiae or L5U1 C. glabrata cells transformed with the pGREG576-CgFLR1 and pGREG576-CgFLR2 or pGREG576-MTI-CgFLR1 and pGREG576-MTI-CgFLR2 plasmids, respectively. These cells express the CgFlr1_GFP or CgFlr2_GFP fusion proteins, whose localization may be determined using fluorescence microscopy. S. cerevisiae cell suspensions were prepared by cultivation in BM-U medium, containing 0.5% glucose and 0.1% galactose, at 30°C, with orbital shaking (250 rpm), until a standard culture OD600nm = 0.4 ± 0.04 was reached. At this point cells were transferred to the same medium containing 0.1% glucose and 1% galactose, to induce protein expression. C. glabrata cell suspensions were prepared in BM-U medium, until a standard culture OD600nm = 0.4 ± 0.04 was reached, and transferred to the same medium supplemented with 50 μM CuSO4 (Sigma), to induce the expression of the fusion protein. After 5 h of incubation, the distribution of CgFlr1_GFP or CgFlr2_GFP fusion proteins in S. cerevisiae or in C. glabrata living cells was detected by fluorescence microscopy in a Zeiss Axioplan microscope (Carl Zeiss MicroImaging), using excitation and emission wavelength of 395 and 509 nm, respectively. Fluorescence images were captured using a cooled CCD camera (Cool SNAPFX, Roper Scientific Photometrics).
Antifungal susceptibility assays
The susceptibility of the parental strain KUE100 toward toxic concentrations of the selected drugs was compared to that of the deletion mutants KUE100_Δcgflr1 and KUE100_Δcgflr2 by spot assays, using the steps described elsewhere (Costa et al., 2013b). The ability of CgFLR1 and CgFLR2 gene expression to increase wild-type resistance to the tested chemical stresses was also examined in the URA3- strain L5U1 C. glabrata strain, using the pGREG576_MTI_CgFLR1 and pGREG576_MTI_CgFLR2 centromeric plasmids. Additionally, the effect of CgFLR1, and CgFLR2 expression in S. cerevisiae BY4741 wild-type strain and BY4741_Δflr1 single deletion mutant was also carried out as described elsewhere (Costa et al., 2013b). The tested drugs included the following compounds, used in the specified concentration ranges: the azole antifungal drugs ketoconazole (10–60 mg/L), fluconazole (75–250 mg/L), miconazole (0.10–0.50 mg/L), tioconazole (0.2–0.9 mg/L), itraconazole (0.1–20 mg/L), and clotrimazole (1–15 mg/L), the polyene antifungal drug amphotericin B (0.10–0.30 mg/L), the fluoropyrimidine 5-flucytosine (0.01–4 mg/L) and the pesticide mancozeb (0.5–2.5 mg/L) (all from Sigma).
Drug accumulation assays
The internal accumulation of flucytosine or clotrimazole was determined by calculating the ratio between the radiolabeled compound measured within the yeast cells and in the external medium (Intracellular/Extracellular). The parental strain KUE100 and the mutant strains KUE100_Δcgflr1 and KUE100_Δcgflr2 were grown in BM medium until mid-exponential phase and harvested by filtration. Cells were washed and resuspended in BM medium, to obtain dense cell suspensions (OD600nm = 0.5 ± 0.1, equivalent to ~ 1.57 mg (dry weight) mL−1). Readily, 0.1 μM of 3H- flucytosine or 3H- clotrimazole (American Radiolabelled Chemicals; 1 mCi/ml) and sub-inhibitory concentrations of the corresponding cold drug were added to the cell suspensions. Incubation proceeded for an additional period of 30 min. The intracellular accumulation of labeled antifungal was followed by filtering 200 μl of cell suspension, at adequate time intervals, through pre-wetted glass microfiber filters (Whatman GF/C). The filters were washed with ice-cold TM buffer and the radioactivity measured in a Beckman LS 5000TD scintillation counter. Extracellular 3H-drug was estimated, by radioactivity assessment of 50 μl of the supernatant. Non-specific 3H-drug adsorption to the filters and to the cells (<5% of the total radioactivity) was assessed and taken into consideration. To calculate the intracellular concentration of labeled antifungal drug, the internal cell volume (Vi) of the exponential cells, grown in the absence of drug and used for accumulation assays, was considered constant and equal to 2.5 μl (mg dry weight)−1 (Rosa and Sá-Correia, 1996). Statistical analysis of the results was performed using analysis of variance, and differences were considered significant for P < 0.05.
CgFLR1 and CgFLR2 expression measurements
The levels of CgFLR1 and CgFLR2 transcripts were assessed by real-time PCR, using the approach described before (Costa et al., 2013b). Synthesis of cDNA for real time RT-PCR experiments, from total RNA samples, was performed using the Multiscribe™ reverse transcriptase kit (Applied Biosystems) and the 7500 RT-PCR Thermal Cycler Block (Applied Biosystems), following the manufacturer's instructions. The quantity of cDNA for the following reactions was kept around 10 ng. The subsequent RT-PCR step was carried out using SYBR® Green reagents. Primers for the amplification of the CgFLR1, CgFLR2, and CgACT1 cDNA were designed using Primer Express Software (Applied Biosystems) and are—TCTTATTCACGATGCTACAAATTGG -3′ and 5′-GAATCACAAGGCCAGCAAAGTT -3′; 5′-GCAGCGGCATTCCCATTAT -3′ and 5′-CGGGATACTTTTTTGTGCTCAAT -3′; and 5′-AGAGCCGTCTTCCCTTCCAT -3′ and 5′-TTGACCCATACCGACCATGA -3′, respectively. The RT-PCR reaction was carried out using a thermal cycler block (7500 Real-Time PCR System—Applied Biosystems). Default parameters established by the manufacturer were used and fluorescence detected by the instrument and registered in an amplification plot (7500 System SDS Software—Applied Biosystems). The CgACT1 mRNA level was used as an internal control. The relative values obtained for the wild-type strain in control conditions were set as 1 and the remaining values are presented relative to that control. To avoid false positive signals, the absence of non-specific amplification with the chosen primers was confirmed by the generation of a dissociation curve for each pair of primers. Statistical analysis of the results was performed using analysis of variance, and differences were considered significant for P < 0.05.
Results
Membrane proteome-wide changes occurring in response to flucytosine in C. glabrata
Given the importance of membrane proteins as a first line of defense against external stress agents, the membrane proteome of C. glabrata cells exposed to flucytosine-induced stress was compared to that of unstressed cells. Details on the protein quantification can be assessed at the Mass spectrometry Interactive Virtual Environment (MassIVE) repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp; MassIVE ID: MSV000079209). Annotated spectra for single peptide identification for each protein in the membrane-associated proteome is provided in MS-viewer (http://prospector2.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer; Search keys: qrvm65goix; xffxn22szm; tm0y9x0hqd) (Pais et al., 2016).
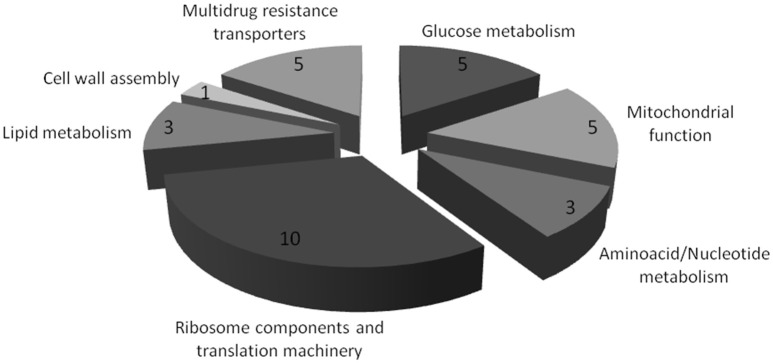
Among the identified membrane-associated proteins, 32 were found to exhibit more than 1.5-fold increased or decreased concentrations in C. glabrata cells exposed to inhibitory concentrations of flucytosine, when compared to the same cells growing in the absence of stress. Within these proteins, 21 were found to be down-regulated and 11 up-regulated in flucytosine challenged cells. Categorization of these proteins, based on their function predicted by homology to their S. cerevisiae homologs, enabled their clustering into seven groups: Glucose metabolism; Mitochondrial function; Amino acid/Nucleotide metabolism; Ribosome components and translation machinery; Lipid metabolism; Cell wall assembly; Multidrug Resistance transporters (Figure 1; Table 1).
Figure 1.
Major functional groups found to have significant expression changes in the C. glabrata membrane-enriched proteome upon exposure to 5-flucytosine. Proteins with significant expression changes include glucose metabolism (5 proteins), mitochondrial function (5 proteins), aminoacid/nucleotide metabolism (3 proteins), ribosome components and translation machinery (10 proteins), lipid metabolism (3 proteins), cell wall assembly (1 protein), and multidrug resistance transporters (5 proteins).
Table 1.
Set of 32 proteins found to have significant expression changes in C. glabrata wild-type cells in the presence of flucytosine and correspondent fold changes in Δcgpdr1 mutant cells upon exposure to the drug.
| C. glabrata protein (ORF) name | S. cerevisiae homolog | Description of the function of the C. glabrata protein or of its S. cerevisiae homolog | Wild-type fold change (upon 5-flucytosine stress) | Δcgpdr1 fold change (upon 5-flucytosine stress) |
|---|---|---|---|---|
| GLUCOSE METABOLISM | ||||
| CgPDC1 (CAGL0M07920g) | PDC1 | Pyruvate decarboxylase, involved in pyruvate metabolism | 0.54 | 0.39 |
| CAGL0L01485g | GSF2 | Putative protein of the ER membrane involved in hexose transporter secretion | 0.60 | 0.43 |
| CgPGK1 (CAGL0L07722g) | PGK1 | Putative 3-phosphoglycerate kinase | 0.20 | 0.63 |
| CgFBA1 (CAGL0L02497g) | FBA1 | Fructose-bisphosphate aldolase | 0.58 | 0.47 |
| CAGL0G06138g | YCK1 | S. cerevisiae ortholog encodes a palmitoylated plasma membrane-bound casein kinase I isoform; functions in morphogenesis, endocytic trafficking, and glucose sensing | 0.44 | 0.90* |
| MITOCHONDRIAL FUNCTION | ||||
| CgRIP1 (CAGL0I03190g) | RIP1 | Putative ubiquinol-cytochrome C reductase iron-sulfur protein | 1.68 | 0.28 |
| CAGL0F04213g | AAC2 | S. cerevisiae ortholog encodes a major ADP/ATP carrier of the mitochondrial inner membrane | 0.64 | 1.00* |
| CAGL0C02695g | MDM10 | Ortholog(s) have role in establishment of mitochondrion localization, mitochondrial outer membrane translocase complex assembly, phospholipid transport, protein import into mitochondrial outer membrane | 0.51 | 0.91* |
| CAGL0L06490g | PHB2 | Ortholog(s) have role in mitochondrion inheritance, negative regulation of proteolysis, protein folding and replicative cell aging | 0.63 | 0.85* |
| CAGL0M09713g | YIM1 | Putative protein involved in DNA damage response | 0.24 | 0.48 |
| AMINO ACID / NUCLEOTIDE METABOLISM | ||||
| CgILV5 (CAGL0B03047g) | ILV5 | Ketol-acid reducto-isomerase | 0.50 | 0.70* |
| CgURA3 (CAGL0I03080g) | URA3 | Orotidine 5'-phosphate decarboxylase, catalyzes a step in pyrimidine biosynthesis; converts 5-FOA into 5-fluorouracil, a toxic compound | 0.44 | 2.92 |
| CAGL0M12881g | URA1 | Ortholog(s) have dihydroorotate oxidase (fumarate) activity, role in 'de novo' pyrimidine nucleobase biosynthetic process | 0.26 | 2.62 |
| RIBOSOME COMPONENTS AND TRANSLATION MACHINERY | ||||
| CAGL0L03846g | DBP2 | Ortholog(s) have RNA-dependent ATPase activity and role in mRNA catabolic process, nonsense-mediated decay, rRNA processing | 3.94 | 2.15 |
| CAGL0E03938g | RPL4B | S. cerevisiae ortholog encodes a ribosomal 60S subunit protein L13B | 1.56 | 1.30* |
| CAGL0K07414g | RPL20B | S. cerevisiae ortholog encodes a ribosomal 60S subunit protein L20A | 1.70 | 0.97* |
| CAGL0J03234g | RPS24B | Ortholog(s) have role in maturation of SSU-rRNA from tricistronic rRNA transcript | 1.52 | 1.05* |
| CAGL0K01859g | NOP1 | Ortholog(s) have mRNA binding, rRNA methyltransferase activity and role in box C/D snoRNA 3'-end processing, rRNA methylation | 1.64 | 0.86* |
| CAGL0I00792g | RPS16A | Ortholog(s) have role in maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) and 90S preribosome | 1.97 | 1.27* |
| CAGL0G01078g | RPL26A | Ortholog(s) have RNA binding, structural constituent of ribosome activity, role in cytoplasmic translation and cytosolic large ribosomal subunit | 1.78 | 1.37* |
| CAGL0E02013g | RPL18A | S. cerevisiae ortholog encodes a ribosomal 60S subunit protein L18A | 0.59 | 0.48 |
| CAGL0L06886g | RPL13B | S. cerevisiae ortholog encodes a ribosomal 60S subunit protein L13B | 0.62 | 0.96* |
| CAGL0A03278g | RPL19A | S. cerevisiae ortholog encodes a ribosomal 60S subunit protein L19A | 0.33 | 0.55 |
| LIPID AND CELL WALL METABOLISM | ||||
| CAGL0L03828g | CYB5 | Ortholog(s) have electron carrier activity, role in ergosterol biosynthetic process | 2.14 | 0.98* |
| CAGL0E03201g | CHO2 | Ortholog(s) have phosphatidylethanolamine N-methyltransferase activity, role in phosphatidylcholine biosynthetic process | 1.56 | 1.56 |
| CAGL0M08206g | YJL171c | S. cerevisiae ortholog encodes a GPI-anchored cell wall protein of unknown function; induced in response to cell wall damage | 0.59 | 0.5 |
| CgHFD1 (CAGL0K03509g) | HFD1 | Putative mitochondrial fatty aldehyde dehydrogenase | 0.29 | 0.24 |
| MULTIDRUG RESISTANCE TRANSPORTERS | ||||
| CgFLR1 (CAGL0H06017g) | FLR1 | Multidrug transporter of the major facilitator superfamily; | 2.08 | 1.89** |
| CgSNQ2 (CAGL0I04862g) | SNQ2 | Predicted plasma membrane ATP-binding cassette (ABC) transporter, putative transporter involved in multidrug resistance | 0.61 | 0.75* |
| CgCDR1 (CAGL0M01760g) | PDR5 | Multidrug transporter of ATP-binding cassette (ABC) superfamily, involved in resistance to azoles | 0.30 | 0.1 |
| CgYOR1 (CAGL0G00242g) | YOR1 | Putative ABC transporter involved in multidrug efflux | 0.51 | 0.44 |
| CgQDR2 (CAGL0G08624g) | QDR2 | Drug:H+ antiporter of the Major Facilitator Superfamily, confers imidazole drug resistance | 0.57 | 0.31 |
The name of the proteins whose expression change was found to vary more than 1.5-fold in the Δcgpdr1 mutant when compared to the wild-type is underlined. Protein description and clustering was obtained from the Candida Genome Database (www.candidagenome.org) or, when completely uncharacterized, based on the role of their predicted S. cerevisiae homologs, obtained from the Yeast Genome Database (www.yeastgenome.org).
Fold change value outside of the chosen cut-off intervals (0.67 < fold change < 1.5);
Fold change quantification considered as not statistically significant (p > 0.05).
The largest functional group identified in the 5-FC membrane proteome response, including a third of the proteins with altered content, is related to RNA metabolism. The expression of seven proteins involved in ribosome biogenesis and translation was found to increase in flucytosine stressed cells, whereas three proteins of the same category were found to be down-regulated in these conditions. It is difficult to say what the exact consequences of the altered expression of each of them individually may be. Additionally, the concentration of several proteins related to glucose metabolism and mitochondrial function related proteins was found to decrease in the presence of flucytosine. The expression of one amino acid biosynthetic protein, Ilv5, and two pyrimidine biosynthetic proteins, Ura1, and Ura3, was also found to decrease in cells exposed to this antifungal agent. Interestingly, Ura3 is responsible for the conversion of 5-FOA into 5-fluorouracil, a key step in the conversion of flucytosine into its toxic sub-products. The repressed expression of the Ura proteins may confer an advantage to flucytosine stressed cells as it may delay the conversion of this pro-drug into its toxic subproducts. Finally, a group of five multidrug transporters was found to exhibit altered levels of expression in flucytosine stressed cells. Four of them, previously implicated in azole drug resistance (Sanglard et al., 1999; Vermitsky et al., 2006; Torelli et al., 2008; Costa et al., 2013b), were actually found to be down-regulated, while the fifth, CgFlr1, was found to be more than 2-fold up-regulated upon C. glabrata exposure to flucytosine.
Effect of CgPDR1 deletion in the membrane proteome-wide changes occurring in response to flucytosine in C. glabrata
To assess the possible involvement of the transcription factor Pdr1 in the resistance to 5-flucytosine, the growth of the C. glabrata strains 66032u and 66032u_Δcgpdr1 (Vermitsky and Edlind, 2004) was compared in solid medium containing inhibitory concentrations of flucytosine, clotrimazole and fluconazole (Figure 2). CgPDR1 deletion fully abrogates growth in the presence of the tested azole drug concentrations, as expected based on numerous reports on the pivotal role of this transcription factor in azole drug resistance. Interestingly, the Δcgpdr1 deletion mutant was also found to display higher susceptibility to the antifungal drug 5-FC than the wild-type parental strain, suggesting that it plays a role in the resistance to this drug as well. Based on this result, the analysis of the membrane-enriched fraction of the C. glabrata proteome obtained from cells exposed to 5-flucytosine in the absence of the transcription factor CgPdr1 was assessed and compared to that of the C. glabrata wild-type cells exposed to 5-flucytosine. Considering an at least 1.5-fold difference in protein expression activation in wild-type vs. Δcgpdr1 cells exposed to 5-flucytosine, nine proteins were found to be repressed by CgPdr1; while eight proteins were found to be activated by CgPdr1 (Table 1). The PDRE loci BCCRYYRGD, TCCRYGGA (Tsai et al., 2010), TCCACGGA, and HYCCGTGGR (Paul et al., 2014), were searched for in the promoters of the referred genes using the PathoYeastract database (Monteiro et al., 2016). Interestingly, considering these 17 proteins, at least one CgPdr1-binding site is found in the promoter regions of nine genes, suggesting the action of CgPdr1 in their expression may be direct. For the remaining 15 proteins, no statistically significant change could be detected in the current experiment. The proteins whose expression was found to be higher in the wild-type strain than in the Δcgpdr1 deletion mutant include the multidrug transporters CgCdr1 and CgQdr2, but also two proteins related to mitochondrial function, CgCyb5 (ORF CAGL0L03828g) and CgRip1 (ORF CAGL0I03190g), and four proteins involved in RNA metabolism, CgDbp2 (ORF CAGL0L03846g), CgNop1 (ORF CAGL0K01859g), CgRpl20B (ORF CAGL0K07414g), and CgRps16A (ORF CAGL0I00792g).
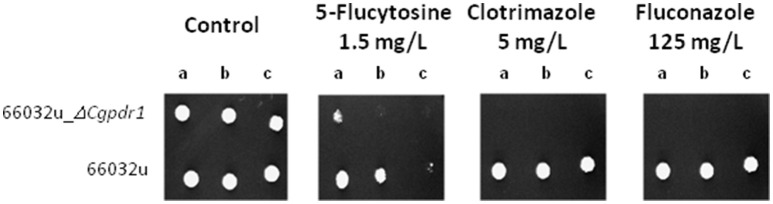
Figure 2.
CgPdr1 confers resistance to flucytosine in C. glabrata cells. Comparison of the susceptibility to inhibitory concentrations of flucytosine, clotrimazole and fluconazole, at the indicated concentrations, of the C. glabrata 66032u and 66032u_Δcgpdr1 strains, in BM plates by spot assays. The inocula were prepared as described under “Materials and Methods.” Cell suspensions used to prepare the spots were 1:5 (b) and 1:25 (c) dilutions of the cells suspension used in (a). The displayed images are representative of at least three independent experiments.
CgFlr1 and CgFlr2 expression confers resistance to other chemical stress inducers
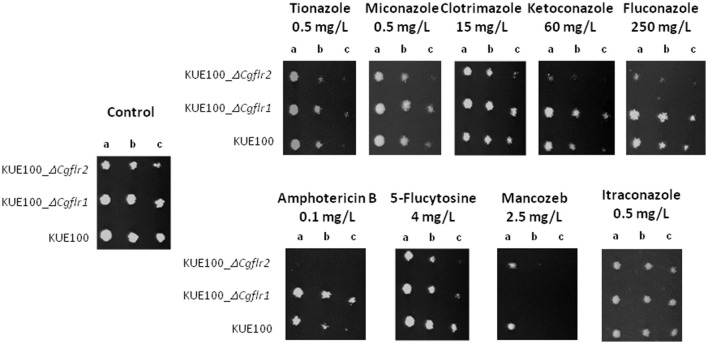
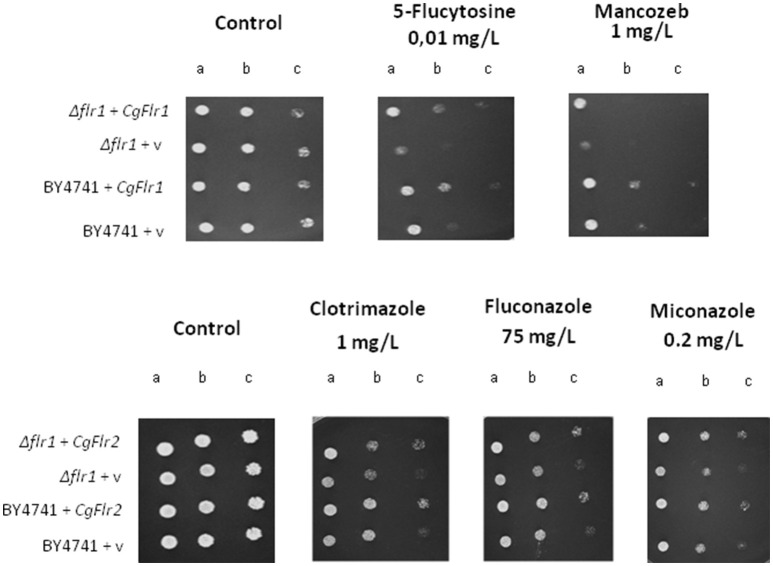
The involvement of CgFlr1 and CgFlr2 in antifungal drug resistance was evaluated, through susceptibility assays, considering a total of nine antifungal drugs of four different families and one agricultural fungicide mancozeb. The results obtained by screening the susceptibility phenotypes of Δcgflr1 and Δcgflr2 mutants, when compared to the wild-type strain, reveal that CgFLR1 confers resistance to mancozeb, whereas CgFLR2 confers resistance to azoles and amphotericin B (Figure 3).
Figure 3.
CgFlr1 and CgFlr2 confer resistance to flucytosine in C. glabrata cells. Comparison of the susceptibility to inhibitory concentrations of several chemical stress inducers, at the indicated concentrations, of the C. glabrata KUE100, KUE100_Δcgflr1 and KUE100_Δcgflr2 strains, in BM plates by spot assays. The inocula were prepared as described under “Materials and Methods.” Cell suspensions used to prepare the spots were 1:5 (b) and 1:25 (c) dilutions of the cells suspension used in (a). The displayed images are representative of at least three independent experiments.
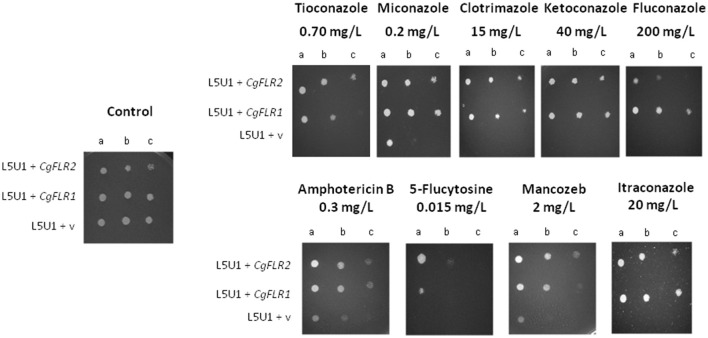
Both CgFLR1 and CgFLR2 were found to confer resistance to flucytosine, although the effect of CgFLR2 is stronger (Figure 3). The expression of CgFLR1-GFP or CgFLR2-GFP in the L5U1 C. glabrata wild-type strain, as verified by anti-GFP western analysis (Figure S1), was concordantly found to increase C. glabrata natural resistance toward the antifungal drugs to which the deletion of each gene was found to lead to a susceptibility phenotype (Figure 4).
Figure 4.
CgFlr1 and CgFlr2 expression increases flucytosine resistance in C. glabrata cells. Comparison of the susceptibility to inhibitory concentrations of several chemical stress inducers, at the indicated concentrations, of the C. glabrata L5U1 strain, harboring the pGREG576 cloning vector (v) or the pGREG576_MTI_CgFLR1 or pGREG576_MTI_CgFLR2 plasmids, in BM-U plates (50 μM CuSO4 supplemented) by spot assays. The inocula were prepared as described under “Materials and Methods.” Cell suspensions used to prepare the spots were 1:5 (b) and 1:25 (c) dilutions of the cells suspension used in (a). The displayed images are representative of at least three independent experiments.
Using S. cerevisiae as a heterologous expression system, the effect of CgFLR1 and CgFLR2 expression on yeast drug resistance was further investigated. When expressed in S. cerevisiae, the CgFLR1 and CgFLR2 genes were found to rescue the susceptibility phenotype exhibited by the S. cerevisiae Δflr1 mutant against flucytosine and mancozeb, and azole drugs, respectively (Figure 5).
Figure 5.
CgFlr1 and CgFlr2 confer resistance to antifungal drugs when heterologously expressed in S. cerevisiae cells. Comparison of the susceptibility to inhibitory concentrations of several chemical stress inducers, at the indicated concentrations, of the S. cerevisiae BY4741 and BY4741_Δflr1 strains, harboring the pGREG576 cloning vector (v) or the pGREG576_CgFLR1 of pGREG576_CgFLR2 plasmids, in BM-U plates by spot assays. The inocula were prepared as described under “Materials and Methods.” Cell suspensions used to prepare the spots were 1:5 (b) and 1:25 (c) dilutions of the cells suspension used in (a). The displayed images are representative of at least three independent experiments.
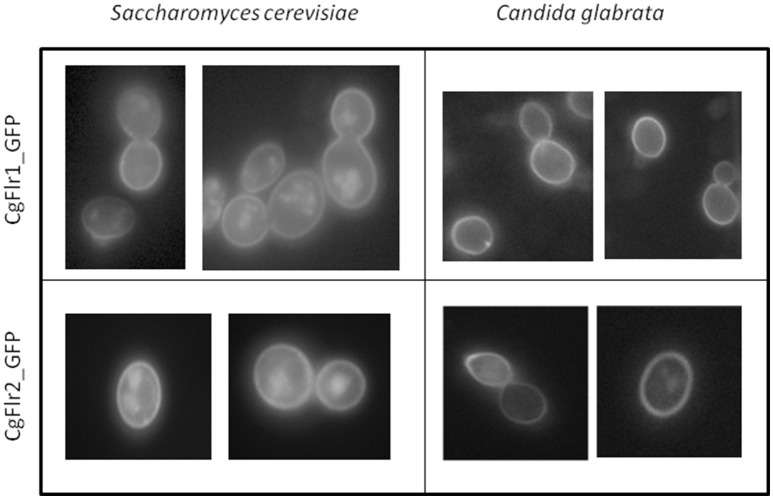
CgFlr1 and CgFlr2 are localized to the plasma membrane
Upon expression induction, C. glabrata cells harboring the pGREG576_MTI_CgFLR1 and pGREG576_MTI_CgFLR2 plasmids were inspected by fluorescence microscopy. The CgFlr1_GFP and CgFlr2_GFP fusion proteins were found to be localized to the cell periphery (Figure 6). In a similar approach, CgFlr1_GFP and CgFlr2_GFP were found to be localized to the cell periphery when expressed heterologously in S. cerevisiae (Figure 6). These results strongly suggest plasma membrane localization for both CgFlr1 and CgFlr2.
Figure 6.
CgFlr1 and CgFlr2 are plasma membrane proteins. Fluorescence of exponential phase BY4741 S. cerevisiae and L5U1 C. glabrata cells, harboring the expression plasmids pGREG576_CgFLR1 and pGREG576_CgFLR2 or pGREG576_MTI_CgFLR1, and pGREG576_MTI_CgFLR2, after galactose or copper-induced recombinant protein production, respectively.
CgFlr1 and CgFlr2 reduce the intracellular accumulation of antifungal drugs in C. glabrata
Consistent with the observed susceptibility phenotypes, Δcgflr1 and Δcgflr2 deletion mutants were found to accumulate 2-fold more radiolabeled flucytosine than the KUE100 parental strain (Figures 7A,B).
Figure 7.
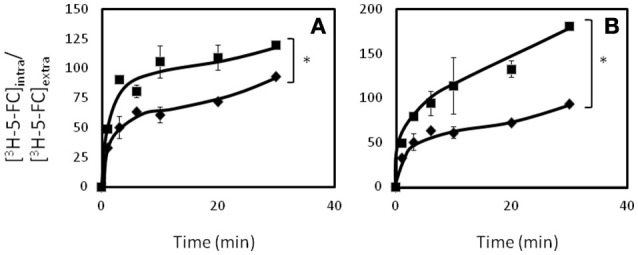
CgFlr1 and CgFlr2 expression decreases the intracellular accumulation of 3H-flucytosine. Time-course accumulation of radiolabeled 3H-flucytosine in KUE100 wild-type (♦) and KUE100_Δcgflr1 (■) (A) and KUE100 (♦) and KUE100_Δcgflr2 (■) (B) strains, during cultivation in BM liquid medium in the presence of sub-lethal concentrations of unlabeled flucytosine. Accumulation values are the average of at least three independent experiments. Errors bars represent the corresponding standard deviations. *p < 0.05.
Additionally, Δcgflr2 mutant cells were found to accumulate around 8-fold more radiolabelled clotrimazole than the parental strain (Figure 8). These results strongly suggest that CgFlr1 and CgFlr2 activities increase C. glabrata resistance toward flucytosine, and in the case of CgFlr2 toward azoles, by reducing their accumulation in yeast cells.
Figure 8.
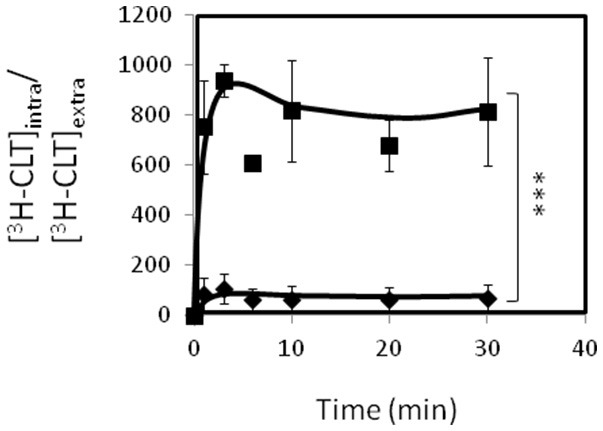
CgFlr2 expression decreases the intracellular accumulation of 3H-clotrimazole. Time-course accumulation of radiolabeled 3H-clotrimazole in KUE100 wild-type (♦) and KUE100_Δcgflr2 (■) strains, during cultivation in BM liquid medium in the presence of sub-lethal concentrations of unlabeled clotrimazole. Accumulation values are the average of at least three independent experiments. Errors bars represent the corresponding standard deviations. ***p < 0.001.
CgFLR1 and CgFLR2 transcript levels are up-regulated under antifungal stress, their basal expression being controlled by CgPdr1 and CgYap1 transcription factors
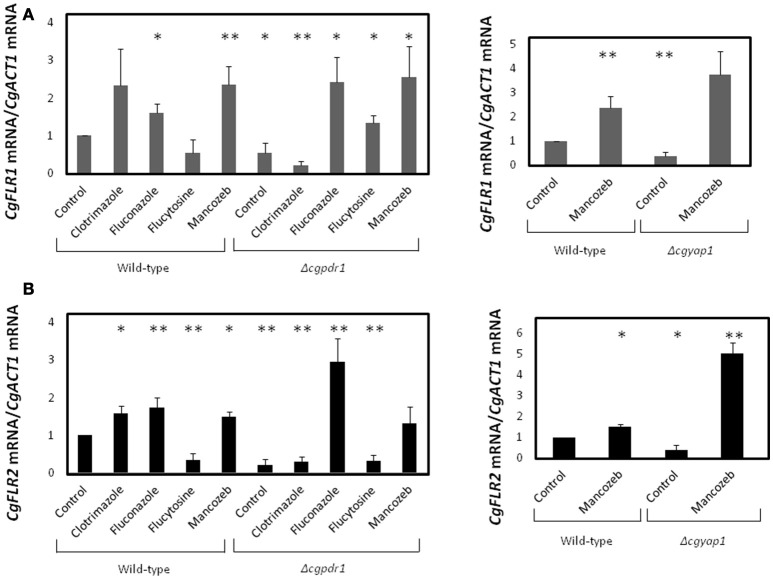
In order to evaluate whether or not the expression of CgFLR1 and CgFLR2 is affected upon drug exposure, quantitative RT-PCR was used to study the effect of flucytosine, clotrimazole, and mancozeb stress exposure in the transcript levels of these genes. No up-regulation of CgFLR1 or CgFLR2 genes could be detected in wild-type cells upon flucytosine exposure. Interestingly, however, CgFLR1 was found to be up-regulated in mancozeb exposed cells, whereas both CgFLR1 and CgFLR2 were found to be up-regulated upon exposure to clotrimazole or fluconazole stress, which is consistent with the role of CgFLR1 and CgFLR2 in mancozeb and azole drug resistance, respectively (Figures 9A,B).
Figure 9.
CgFLR1 and CgFLR2 transcriptional control. Comparison of the variation of the CgFLR1 (A) and CgFLR2 (B) transcript levels in the 66032u C. glabrata wild-type strain and in the derived 66032u_Δcgpdr1 deletion mutant; and in the 84u C. glabrata wild-type strain and in the derived 84u_Δcgyap1 deletion mutant, before and after 1 h of exposure to 60 mg/L clotrimazole, 80 mg/L fluconazole, 8 mg/L flucytosine and 20 mg/L mancozeb. The presented transcript levels were obtained by quantitative RT-PCR and are relative CgFLR1/CgACT1 or CgFLR2/CgACT1 mRNA, relative to the values registered in the 66032 or 84u parental strains in control conditions. The indicated values are averages of at least three independent experiments. Error bars represent the corresponding standard deviations. *p < 0.05; **p < 0.01.
Additionally, given the fact that the transcription factor CgPdr1 is the master regulator of azole drug resistance (Tsai et al., 2006) and CgYap1 had been previously linked to the control of CgFlr1 expression (Chen et al., 2007), the effect of CgYap1 and CgPdr1 deletion in the control of the expression of CgFLR1 and CgFLR2 was further evaluated. CgFLR1 and CgFLR2 up-regulation, registered under clotrimazole exposure—but not under fluconazole stress —, was found to be abrogated in the absence of CgPDR1 (Figure 9B), while CgPdr1 and CgYap1 were found to control the basal expression of both CgFLR1 and CgFLR2 (Figures 9A,B). As expected, based on the proteomics data, none of the genes was found to be controlled by CgPdr1 in flucytosine exposed cells. Unexpectedly, the up-regulation of CgFLR1, and CgFLR2 registered under mancozeb stress was found not to be controlled by CgYap1. Indeed, in Δcgyap1 cells the mancozeb-induced up-regulation of these genes was found to be even stronger than that registered in the wild-type strain (Figures 9A,B).
Discussion
5-Flucytosine has fallen into disuse due to the rapid acquisition of resistance by fungal pathogens and to its moderate toxicity in humans, limiting the administration of higher dosages. The identification of the mechanisms underlying these phenomena is, thus, crucial to maintain the use of 5-FC as a therapeutic option. In this work, the changes occurring at the membrane proteome level in C. glabrata cells exposed to 5-FC were analyzed, highlighting new mechanisms of resistance to this antifungal drug.
One of the most interesting aspects of this work concerns the identification of CgPdr1 transcription factor, a major factor of resistance to azoles (Tsai et al., 2006), as a determinant of 5-FC resistance. The deletion of CgPdr1 was consistently found to decrease the activation of about 50% of the membrane proteins found to be up-regulated in response to 5-FC. Among the proteins whose expression was seen to be affect by CgPdr1 deletion are the multidrug efflux transporter encoding genes CgCDR1 and CgQDR2, but also CgRIP1, encoding a putative ubiquinol-cytochrome C reductase iron-sulfur protein, which were previously found to be controlled by this transcription factor (Tsai et al., 2006; Vermitsky et al., 2006; Ferrari et al., 2011; Costa et al., 2013b; Pais et al., 2016). New putative targets of CgPdr1 up-regulated in the context of 5-FC response include four proteins involved in RNA metabolism, CgDbp2, CgNop1, CgRpl20B, and CgRps16A. Interestingly, the traditional targets of CgPdr1, including the ABC drug efflux pumps CgCdr1, CgYor1, and CgSnq2 are repressed in response to 5-FC, which suggests that, although being active in the response to azoles and to 5-FC, the action of Pdr1 at the level of transcriptional control appears to be different. It will be interesting to test whether this differential outcome of CgPdr1 activity is linked to differences in terms of the conformation of this transcription factor, whose activation is known to occur by the direct binding to the drug molecule (Thakur et al., 2008). To gain a full view on the extension of the participation of CgPdr1 in 5-FC response, however, it would be necessary to conduct a transcriptomics study.
A large proportion of the 5-FC response was found to be related to RNA and protein metabolism. Interestingly, an increased expression of some ribosome and translation associated proteins was observed, which may be related to the specific mechanism of action of 5-FC. It is thus possible to assume that the RNA- and protein-metabolism-related genes identified herein as responding to 5-FC challenge may be involved in counteracting its primary toxic action. Previous analyses of the transcriptome-wide S. cerevisiae (Zhang et al., 2002) or Candida albicans (Liu et al., 2005) response to 5-FC, also highlighted the relevance of RNA metabolism in the response to this antifungal drug. Indeed, Liu et al. found an up-regulation of several genes involved in RNA metabolism and translation (Liu et al., 2005), while Zhang and co-workers showed a down regulation of a few ribosomal protein encoding genes in these conditions (Zhang et al., 2002). It is also in agreement with a previous chemogenomic analysis of the determinants of 5-FC resistance in the model yeast S. cerevisiae, in which about one fourth of the determinants of resistance to this drug were found to be related to RNA and protein metabolism (Costa et al., 2015). It appears that C. glabrata cells try to compensate, with the increased expression of translation associated proteins, the detrimental effect that 5-FC exerts in this process.
Another interesting feature of the proteomics response includes the down-regulation of two nucleotide biosynthesis related proteins, CgUra1 and CgUra3. CgUra1 catalyses the synthesis of ororate from dihydroorotate, which is, then converted to oritidine-5-phosphate. CgUra3 catalyses the next step of conversion of oritidine-5-phosphate to UMP, which is funneled into the production of UDP and UTP, used for RNA synthesis. This same pathway is used to process 5-FC into its toxic products, including 5F-UDP, which upon incorporation in RNA molecules inhibits protein synthesis. It appears, thus, that the cell responds to 5-FC induced stress by decreasing the expression of enzymes required for its conversion to toxic 5-FC products. It is interesting to point out, in this context, that the expression level of CgUra1 and CgUra3 in the Δcgpdr1 deletion mutant is much higher than in wild-type cells. The up-regulation of these proteins in the Δcgpdr1 background may underlie the increased susceptibility to 5-FC exhibited by this deletion mutant, when compared to the wild-type strain.
Among the results obtained from the membrane proteomics analysis, the role of CgFlr1 and of its homolog CgFlr2, in 5-FC response was further analyzed. Although the concentration of CgFlr2 was not found to be increased in the membrane proteome of C. glabrata cells, both CgFlr1, and CgFlr2 were found to confer resistance to 5-FC, apparently due to their role in controlling the levels of 5-FC accumulation within C. glabrata cells. The fact that CgFlr2 is not up-regulated in 5-FC-exposed cells but is required for C. glabrata resistance to this stress, although unexpected, is consistent with the frequent observation that the genes that are required for the resistance to a given stress are not necessarily up-regulated in response to that stress (Teixeira et al., 2011). Interestingly, CgFlr1 was further found to confer resistance to mancozeb, while CgFlr2 was also found to confer resistance to azoles and amphotericin B, placing this transporter at the intersection of multiple antifungal resistance mechanisms. These two transporters constitute, thus, two additional players in the antifungal drug resistance phenomenon. Our group had previously shown that the acquaglyceroporins CgFps1 and CgFps2 (Costa et al., 2015), as well as the DHA transporters CgAqr1 (Costa et al., 2013a) and CgTpo1_1 and CgTpo1_2 (Pais et al., 2016) are determinants of flucytosine resistance as well, suggesting that 5-FC extrusion is an important mechanism of resistance against this antifungal drug and showing this phenomenon to be the consequence of the additive contribution of several players.
The analysis of the expression of the CgFLR1 and CgFLR2 genes further highlighted their importance in the context of drug resistance in C. glabrata. Using the PathoYeastract database (http://pathoyeastract.org/cglabrata/index.php Monteiro et al., 2016), it is possible to verify that the CgFLR1 was found to be up-regulated upon the over-expression of CgPdr1, in control conditions (Noble et al., 2013), or upon benomyl or selenite exposure, in the dependency of CgYap1 (Chen et al., 2007; Lelandais et al., 2008; Merhej et al., 2016). Additionally, CgFLR1 expression was shown to be repressed by the transcription factor Stb5, a negative regulator of azole resistance in C. glabrata (Noble et al., 2013). Chromatine ImmunoPrecipitation (ChIP) assays further showed that Yap7 binds to the CgFLR1 promoter in selenite exposed cells (Merhej et al., 2016). Information on the regulation of CgFLR2 is much scarcer. RNA sequencing data have demonstrated that the predicted zinc cluster transcription factor encoded by ORF CAGL0I07755g, a homolog of the S. cerevisiae Hal9 and of the C. albicans Tac1 transcription factors, is a positive regulator of CgFLR2 in control conditions (Wu et al., 2015). The new data on the regulation of the CgFLR1 and CgFLR2 genes coming from this study shows that both genes are controlled at the transcription level by CgPdr1 in the presence of the azole drug clotrimazole, but not in the presence of fluconazole. The fact that in previous genome-wide expression analysis of fluconazole response in C. glabrata the regulation of CgFLR1 or CgFLR2 by CgPdr1 had not been identified suggests that this effect may be specific to imidazole antifungals, such as clotrimazole, but not to triazole antifungals such as fluconazole. Despite the fact that a previous ChIP experiment probing CgPdr1 targets in ρ0 C. glabrata cells did not pinpoint CgFLR1 or CgFLR2 promoters as targets of CgPdr1 (Paul et al., 2014), a putative CgPdr1 binding site can be found in the promoter region of CgFLR2. The possibility, however, that CgPdr1 may regulate the expression of CgFLR1 and CgFLR2 in clotrimazole stressed cells through direct binding to their promoter regions remains to be established. Given the importance of CgPdr1 in the clinical acquisition of azole drug resistance, these results raise the hypothesis that CgFlr1 and CgFlr2 may be relevant in the clinical context. In a previous transcriptomics analysis of the impact of CgPdr1 GOF mutations in fluconazole resistant isolates, when compared to susceptible ones, no significant changes were detected in terms of the expression of CgFLR1 or CgFLR2 (Tsai et al., 2010). However, it would indeed be interesting to assess whether there is a possible correlation between the expression of these genes and the level of antifungal drug resistance in clinical isolates displaying differential clotrimazole or 5-FC susceptibility phenotypes. Additionally, the fact that the transcript levels of both CgFLR1 and CgFLR2 genes is controlled, at least at the basal level, by CgPdr1 and CgYap1, appears to correlate with the complex regulation of their S. cerevisiae homolog Flr1. The observation that S. cerevisiae FLR1 gene is also strongly up-regulated by the transcription factor Yap1 in mancozeb stressed cells (Teixeira et al., 2008) urged us to check for a similar effect in C. glabrata CgFLR1 and CgFLR2 genes. However, although an increased expression of CgFLR1 and CgFLR2 was indeed registered in C. glabrata cells exposed to mancozeb, this proved to be irrespective of CgYap1 activity, suggesting that the control of the expression of these genes is not fully conserved in C. glabrata. The transcriptional control of ScFlr1 was found to be highly complex, relying on the combined efforts of at least four transcription factors, Yap1, Pdr1, Yrr1, and Rpn4 (Alarco et al., 1997; Brôco et al., 1999; Teixeira et al., 2008). The building of a mathematical model to describe the ScFlr1 regulatory network highlighted that this network is likely to require a fifth, still unidentified, transcription factor to explain the experimental observations, putting forward the hypothesis that its regulation may be even more complex than initially foreseen (Teixeira et al., 2010; Monteiro et al., 2011). It would be interesting to check whether the homologs in C. glabrata of these additional S. cerevisiae transcription factors may also be important in the regulation of these genes, whose control appears to be phylogenetically conserved among these yeast species.
In conclusion, the results described in this study highlight the importance of the DHA transporters from the MFS in antifungal resistance. This work highlights the importance of proteome-wide approaches in the identification of new antifungal resistance mechanisms. The newly identified processes stand out as promising targets for the development of new 5-flucytosine chemosensitizers, which would expectedly allow for the use of decreased therapeutic dosages of 5-flucytosine.
Author contributions
PP and CP conducted most of the experiments and contributed to the writing of the manuscript. CC and MO contributed to development of some of the experimental work. HC and MT conceived the experiments. MT supervised the scientific and experimental design of the work and wrote the manuscript.
Funding
This work was supported by FEDER and “Fundação para a Ciência e a Tecnologia” (FCT) (Contract PTDC/BBB-BIO/4004/2014 and Ph.D. and post-doctoral grants to PP (SFRH/BD/110956/2015) and CC (SFRH/BPD/100863/2014), respectively). Funding received by iBB-Institute for Bioengineering and Biosciences from FCT-Portuguese Foundation for Science and Technology (UID/BIO/04565/2013) and from Programa Operacional Regional de Lisboa 2020 (Project N. 007317) is acknowledged.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thomas Edlind, from the Department of Microbiology and Immunology, Drexel University, College of Medicine, Philadelphia, USA, is acknowledged for kindly providing the 66032u derived strains. John Bennett, from the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, USA, is acknowledged for kindly providing the L5U1 and 84u strains.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02045/full#supplementary-material
CgFlr1 and CgFlr2 are expressed in C. glabrata cells harboring the pGREG576_MT1_CgFLR1 and the pGREG576_MT1_CgFLR2 plasmids. Comparison of the level of expression of CgFlr1-GFP or CgFlr2-GFP fusion proteins in L5U1 C. glabrata cells upon exposure to the indicated CuSO4 concentrations, based on anti-GFP immuno-detection. Cells harboring the pGREG576 cloning vector (control), exposed to the same CuSO4 concentrations were used as a negative control.
References
- Alarco A. M., Balan I., Talibi D., Mainville N., Raymond M. (1997). AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272, 19304–19313. 10.1074/jbc.272.31.19304 [DOI] [PubMed] [Google Scholar]
- Alarco A. M., Raymond M. (1999). The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brôco N., Tenreiro S., Viegas C. A., Sá-Correia I. (1999). FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on Pdr3 transcriptional regulator. Yeast 15, 1595–1608. [DOI] [PubMed] [Google Scholar]
- Chen K. H., Miyazaki T., Tsai H. F., Bennett J. E. (2007). The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 386, 63–72. 10.1016/j.gene.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Costa C., Dias P. J., Sá-Correia I., Teixeira M. C. (2014a). MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front. Physiol. 5:197. 10.3389/fphys.2014.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C., Henriques A., Pires C., Nunes J., Ohno M., Chibana H., et al. (2013a). The dual role of Candida glabrata drug:H+ antiporter CgAqr1 (ORF CAGL0J09944g) in antifungal drug and acetic acid resistance. Front. Microbiol. 4:170. 10.3389/fmicb.2013.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C., Nunes J., Henriques A., Mira N. P., Nakayama H., Chibana H., et al. (2014b). Candida glabrata drug:H+ antiporter CgTpo3 (ORF CAGL0I10384g): role in azole drug resistance and polyamine homeostasis. J. Antimicrob. Chemother. 69, 1767–1776. 10.1093/jac/dku044 [DOI] [PubMed] [Google Scholar]
- Costa C., Pires C., Cabrito T. R., Renaudin A., Ohno M., Chibana H., et al. (2013b). Candida glabrata drug:H+ antiporter CgQdr2 confers imidazole drug resistance, being activated by transcription factor CgPdr1. Antimicrob. Agents Chemother. 57, 3159–3167. 10.1128/AAC.00811-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C., Ponte A., Pais P., Santos R., Cavalheiro M., Yaguchi T., et al. (2015). New Mechanisms of Flucytosine Resistance in C. glabrata Unveiled by a Chemogenomics Analysis in S. cerevisiae. PLoS ONE 10:e0135110. 10.1371/journal.pone.0135110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos S. C., Teixeira M. C., Dias P. J., Sá-Correia I. (2014). MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Front. Physiol. 5:180. 10.3389/fphys.2014.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlind T. D., Katiyar S. K. (2010). Mutational analysis of flucytosine resistance in Candida glabrata. Antimicrob. Agents Chemother. 54, 4733–4738. 10.1128/AAC.00605-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A. (2008). Mechanisms of resistance to antifungal agents: yeasts and filamentous fungi. Rev. Iberoam Micol. 25, 101–106. 10.1016/S1130-1406(08)70027-5 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Sanguinetti M., Torelli R., Posteraro B., Sanglard D. (2011). Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata. PLoS ONE 6:e17589. 10.1371/journal.pone.0017589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel P. L., Jr., Vazquez J. A., Sobel J. D. (1999). Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12, 80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gbelska Y., Krijger J. J., Breunig K. D. (2006). Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res. 6, 345–355. 10.1111/j.1567-1364.2006.00058.x [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A., Rice L. B. (1999). Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12, 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope W. W., Tabernero L., Denning D. W., Anderson M. J. (2004). Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob. Agents Chemother. 48, 4377–4386. 10.1128/AAC.48.11.4377-4386.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Wu C., Schade B., Thomas D. Y., Whiteway M. (2005). Drag&Drop cloning in yeast. Gene 344, 43–51. 10.1016/j.gene.2004.10.016 [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D. P., Lewis R. E. (2002). Antifungal drug resistance of pathogenic fungi. Lancet 359, 1135–1144. 10.1016/S0140-6736(02)08162-X [DOI] [PubMed] [Google Scholar]
- Lelandais G., Tanty V., Geneix C., Etchebest C., Jacq C., Devaux F. (2008). Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol. 9:R164. 10.1186/gb-2008-9-11-r164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. T., Lee R. E., Barker K. S., Wei L., Homayouni R., Rogers P. D. (2005). Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49, 2226–2236. 10.1128/AAC.49.6.2226-2236.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej J., Thiebaut A., Blugeon C., Pouch J., Ali Chaouche Mel A., Camadro J. M., et al. (2016). A network of paralogous stress response transcription factors in the human pathogen Candida glabrata. Front. Microbiol. 7:645. 10.3389/fmicb.2016.00645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N. N., Prasad T., Sharma N., Payasi A., Prasad R., Gupta D. K., et al. (2007). Pathogenicity and drug resistance in Candida albicans and other yeast species. A review. Acta Microbiol. Immunol. Hung 54, 201–235. 10.1556/AMicr.54.2007.3.1 [DOI] [PubMed] [Google Scholar]
- Monteiro P. T., Dias P. J., Ropers D., Oliveira A. L., Sá-Correia I., Freitas A. T. (2011). Qualitative modeling and formal verification of the FLR1 gene mancozeb response in Saccharomyces cerevisiae. IET Syst. Biol. 5, 308–316. 10.1049/iet-syb.2011.0001 [DOI] [PubMed] [Google Scholar]
- Monteiro P. T., Pais P., Costa C., Manna S., Sa-Correia I., Teixeira M. C. (2016). The PathoYeastract database: an information system for the analysis of gene and genomic transcription regulation in pathogenic yeasts. Nucleic Acids Res. 10.1093/nar/gkw817. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J. A., Tsai H. F., Suffis S. D., Su Q., Myers T. G., Bennett J. E. (2013). STB5 is a negative regulator of azole resistance in Candida glabrata. Antimicrob. Agents Chemother. 57, 959–967. 10.1128/AAC.01278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais P., Costa C., Pires C., Shimizu K., Chibana H., Teixeira M. C. (2016). Membrane Proteome-Wide Response to the Antifungal Drug Clotrimazole in Candida glabrata: role of the transcription factor CgPdr1 and the Drug:H+ Antiporters CgTpo1_1 and CgTpo1_2. Mol. Cell. Proteomics 15, 57–72. 10.1074/mcp.M114.045344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papon N., Nöel T., Florent M., Gibot-Leclerc S., Jean D., Chastin C., et al. (2007). Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob. Agents Chemother. 51, 369–371. 10.1128/AAC.00824-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Bair T. B., Moye-Rowley W. S. (2014). Identification of genomic binding sites for Candida glabrata Pdr1 transcription factor in wild-type and rho0 cells. Antimicrob. Agents Chemother. 58, 6904–6912. 10.1128/AAC.03921-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M. F., Sá-Correia I. (1996). Intracellular acidification does not account for inhibition of Saccharomyces cerevisiae growth in the presence of ethanol. FEMS Microbiol. Lett. 135, 271–274. 10.1111/j.1574-6968.1996.tb08000.x [DOI] [PubMed] [Google Scholar]
- Sá-Correia I., dos Santos S. C., Teixeira M. C., Cabrito T. R., Mira N. P. (2009). Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 17, 22–31. 10.1016/j.tim.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., Bille J. (1999). The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43, 2753–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D., Odds F. C. (2002). Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2, 73–85. 10.1016/S1473-3099(02)00181-0 [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Dias P. J., Monteiro P. T., Sala A., Oliveira A. L., Freitas A. T., et al. (2010). Refining current knowledge on the yeast FLR1 regulatory network by combined experimental and computational approaches. Mol. Biosyst. 6, 2471–2481. 10.1039/c004881j [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Dias P. J., Simões T., Sá-Correia I. (2008). Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem. Biophys. Res. Commun. 367, 249–255. 10.1016/j.bbrc.2007.12.056 [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Mira N. P., Sá-Correia I. (2011). A genome-wide perspective on the response and tolerance to food-relevant stresses in Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 22, 150–156. 10.1016/j.copbio.2010.10.011 [DOI] [PubMed] [Google Scholar]
- Thakur J. K., Arthanari H., Yang F., Pan S. J., Fan X., Breger J., et al. (2008). A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609. 10.1038/nature06836 [DOI] [PubMed] [Google Scholar]
- Torelli R., Posteraro B., Ferrari S., La Sorda M., Fadda G., Sanglard D., et al. (2008). The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol. Microbiol. 68, 186–201. 10.1111/j.1365-2958.2008.06143.x [DOI] [PubMed] [Google Scholar]
- Tsai H. F., Krol A. A., Sarti K. E., Bennett J. E. (2006). Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50, 1384–1392. 10.1128/AAC.50.4.1384-1392.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. F., Sammons L. R., Zhang X., Suffis S. D., Su Q., Myers T. G., et al. (2010). Microarray and molecular analysis of the azole resistance mechanism in Candida glabrata oropharyngeal isolates. Antimicrob. Agents Chemother. 54, 3308–3317, 10.1128/AAC.00535-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K., Matsumoto Y., Uno J., Sasamoto K., Sekimizu K., Kinjo Y., et al. (2011). Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS ONE 6:e24759. 10.1371/journal.pone.0024759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K., Uno J., Nakayama H., Sasamoto K., Mikami Y., Chibana H. (2007). Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryotic Cell 6, 1239–1247. 10.1128/EC.00414-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes A., Guchelaar H. J., Dankert J. (2000). Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J. Antimicrob. Chemother. 46, 171–179. 10.1093/jac/46.2.171 [DOI] [PubMed] [Google Scholar]
- Vermitsky J. P., Earhart K. D., Smith W. L., Homayouni R., Edlind T. D., Rogers P. D. (2006). Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61, 704–722. 10.1111/j.1365-2958.2006.05235.x [DOI] [PubMed] [Google Scholar]
- Vermitsky J. P., Edlind T. D. (2004). Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48, 3773–3781. 10.1128/AAC.48.10.3773-3781.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. C. (1997). Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41, 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins D. A., Shimer G. H., Jr., Cottarel G. (2002). A system for deletion and complementation of Candida glabrata genes amenable to high-throughput application. Gene 292, 141–149. 10.1016/S0378-1119(02)00648-0 [DOI] [PubMed] [Google Scholar]
- Wu J., Chen X., Cai L., Tang L., Liu L. (2015). Transcription factors Asg1p and Hal9p regulate pH homeostasis in Candida glabrata. Front. Microbiol. 6:843. 10.3389/fmicb.2015.00843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang Y., Zhou Y., Zhao Y., Cheng J. (2002). Expression profiling of the response of Saccharomyces cerevisiae to 5-fluorocytosine using a DNA microarray. Int. J. Antimicrob. Agents 20, 444–450. 10.1016/S0924-8579(02)00201-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CgFlr1 and CgFlr2 are expressed in C. glabrata cells harboring the pGREG576_MT1_CgFLR1 and the pGREG576_MT1_CgFLR2 plasmids. Comparison of the level of expression of CgFlr1-GFP or CgFlr2-GFP fusion proteins in L5U1 C. glabrata cells upon exposure to the indicated CuSO4 concentrations, based on anti-GFP immuno-detection. Cells harboring the pGREG576 cloning vector (control), exposed to the same CuSO4 concentrations were used as a negative control.