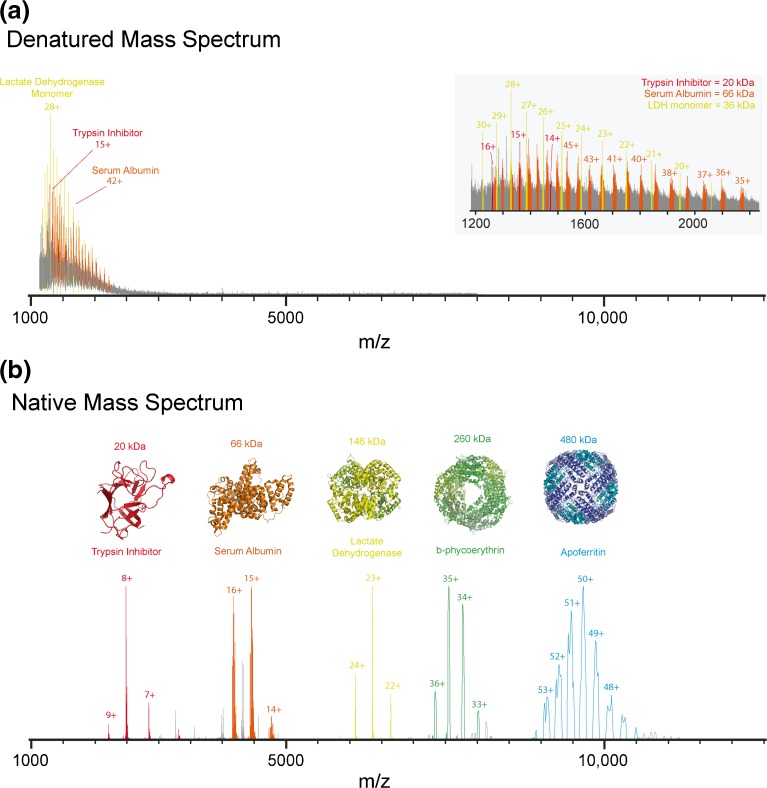
Figure 2.
Comparison of the MS analysis of an identical protein mixture under denatured and native MS conditions. A mixture of proteins, trypsin inhibitor, bovine serum albumin, lactate dehydrogenase, B-phycoerythrin, and apoferritin, commonly used as molecular weight markers in native PAGE, was buffer-exchanged into a water/acetonitrile/formic acid (50/49/1) solution or 100 mM aqueous ammonium acetate pH 7 for denatured MS (a) and native MS (b), respectively. For native MS analysis, the MS parameters on the Orbitrap EMR were optimised specifically for the m/z window of each protein/protein complex and the mass spectra “stitched” together. The proteins corresponding to trypsin inhibitor, bovine serum albumin, lactate dehydrogenase, B-phycoerythrin, and apoferritin are highlighted in red, orange, yellow, green, and blue, respectively. Native MS shows all proteins and protein complexes are widely separated in m/z space, in sharp contrast to denaturing MS where the m/z of the ions corresponding to all the proteins and protein subunits are collapsed in the narrow 1000–2500 m/z range