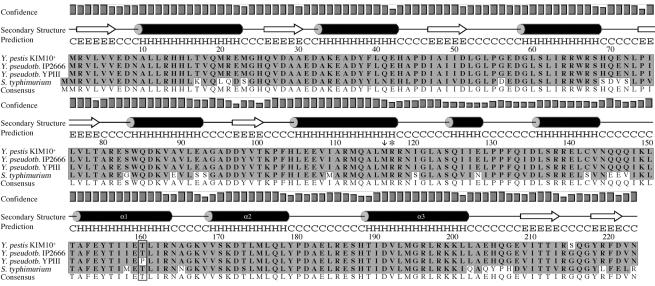
FIG. 3.
Predicted secondary structure of PhoPKIM and ClustalW alignment of PhoP sequences (single-letter code) from Yersinia species and serovar Typhimurium. The secondary structure was predicted by using PSIPRED. H, α-helix; E, β-sheet; C, coil; Y. pseudotb, Y. pseudotuberculosis; S. typhimurium, S. enterica serovar Typhimurium. The confidence levels for secondary structure assignments are indicated by bar graphs. The amino acid sequences of PhoP proteins for KIM10+ (accession no. NP_669111), IP2666 (accession no. AAK54056), and serovar Typhimurium (accession no. RGEBFT) were obtained from GenBank. The sequence of PhoPYPIII was deduced from the nucleotide sequence of the phoP gene from YPIII(p−) (phoPYPIII) obtained in this study. Sequences were aligned by using the ClustalW alignment function in MacVector. The T160P substitution in PhoPYPIII is enclosed in a box. The arrow at position 116 indicates the first codon in phoPYP1II(p+) that is changed (from M to H) due to a frameshift mutation corresponding to a repeat of nucleotides 341 to 345. The asterisk indicates the position of the stop codon in phoPYP1II(p+) which results from the frameshift mutation.