Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is still one of the most important hospital pathogen globally. The multiresistant isolates of the ST239-SCCmecIII lineage are spread over large geographic regions, colonizing and infecting hospital patients in virtually all continents. The balance between fitness (adaptability) and virulence potential is likely to represent an important issue in the clonal shift dynamics leading the success of some specific MRSA clones over another. The accessory gene regulator (agr) is the master quorum sensing system of staphylococci playing a role in the global regulation of key virulence factors. Consequently, agr inactivation in S. aureus may represent a significant mechanism of genetic variability in the adaptation of this healthcare-associated pathogen. We report here the complete genome sequence of the methicillin-resistant S. aureus, isolate HC1335, a variant of the ST239 lineage, which presents a natural insertion of an IS256 transposase element in the agrC gene encoding AgrC histidine kinase receptor.
Keywords: methicillin-resistant Staphylococcus aureus, ST239 lineage, agrC dysfunction, IS256 insertion, complete genome sequence
Introduction
Despite the recent improvements on control infection measures, methicillin-resistant Staphylococcus aureus (MRSA) is still a major health problem concern worldwide (Figueiredo and Ferreira 2014). The isolates of the ST239(CC8)-SCCmecIII lineage—one of the most largely disseminated healthcare-associated MRSA (HA-MRSA)—are endemic in several hospitals and have been described in outbreaks around the world (Li et al. 2012; Figueiredo and Ferreira 2014; Baines et al. 2015). In Brazil, MRSA isolates of a multiresistant S. aureus clone—the Brazilian epidemic clone (BEC) of the lineage ST239-SCCmecIII—are disseminated in hospitals of different regions (Teixeira et al. 1995; Campos et al. 2012; Martins et al. 2014). However, the factors involved in the predominance of a MRSA clone over another remains unclear (Figueiredo and Ferreira 2014). The accessory gene regulator (Agr) is the main S. aureus quorum-sensing system in the control of assorted virulence factors and key mechanisms associated with the pathogenesis of S. aureus infections, including biofilm development (Novick et al. 2000; Ferreira et al. 2012).
The agr locus has two divergent promoters, P2 and P3. P2 promoter guides the transcription of the genes agrBDCA whereas P3 promoter controls the transcription of the main agr effector molecule, the RNAIII. The AgrD protein is processed—to produce a small auto-inducing peptide (AIP)—and secreted by the AgrB protein. In the bacterial cell membrane, AIP binds to and activates the AgrC receptor histidine kinase. The activated AgrC phosphorylates the response regulator AgrA that binds to P2 and P3 promoter regions to activate the transcription of the whole agr locus. RNAIII directly regulates some surface anchored proteins involved in bacterial–host interactions such as protein A (Spa), coagulase (Coa) and fibrinogen-binding protein (Fib), and indirectly modulates—via Rot inactivation—the expression of a number of exoproteins including serine proteases SspA and SspB, α-hemolysin (Hla) and lipase Geh (Novick et al. 2000; Boisset et al. 2007). Thus, the Agr system plays an important role in the severity of the staphylococcal infections (Le and Otto 2015). In addition to RNAIII, the AgrA response regulator can also regulates phenol-soluble modulins (PSM) α and β by directly binding the promoter regions (Queck et al. 2008).
agr-dysfunctional S. aureus isolates have shown increased competitive fitness compared with agr-functional isolates (Paulander et al. 2012). In addition, the bacterial exposure to selected antimicrobials caused increased agr expression and consequent reduction in bacterial fitness (Paulander et al. 2012).
Here we report the complete genome sequence of the MRSA isolate HC1335 (GenBank accession number CP012012.1), a ST239 isolate, presenting a natural agr dysfunction, obtained from nasal colonization of a patient in a home care system, in 2001, as part of a genome project with ST239 isolates collected from infection and colonization cases in Brazil.
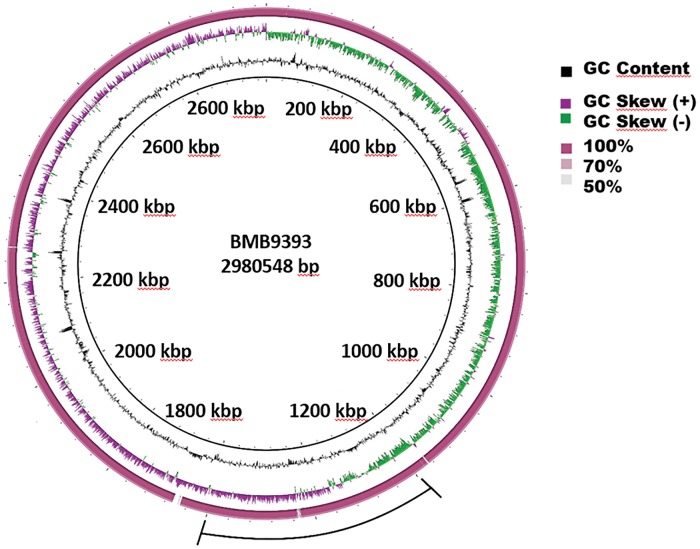
The genome of HC1335 consists of one circular chromosome with 2,976,370 bp and GC content of 32.89% (fig. 1). Functional annotation showed 2,740 protein-coding sequences (CDS)—2,330 of them were assigned to known functions and 410 to unknown categories—and a total of 39 pseudogenes were annotated. In addition, the genome harbors four copies of 16S rRNA, four of 23S rRNA, and five of 5S rRNA, and 57 tRNA genes. We have previously reported the complete genome sequence of an agr-functional ST239 isolate named BMB9393, collected from bloodstream infection in a hospitalized patient (Costa et al. 2013; GenBank acession number CP005289.1). Those two isolates share a core genome with 2,417 coding sequences (CDS). HC1335 genome has 13 exclusive CDS, the vast majority (11) located in a phage region related to phage ϕNM25 at coordinates 1,169,991 to 1,192,826 (supplementary table S1, Supplementary Material online). In addition, HC1335 has an oxide nitric reductase, quinol dependent (qNOR) that lacks the cytochrome c component and uses quinol as the electron donor. This enzyme is present in several pathogens that are not denitrifiers, and the qNOR of HC1335 has 100% amino acid sequence identity with that of S. aureus Z172 (Uniprot: A0A0E1AHM6). The other exclusive CDS is an uncharacterized acyltransferase with 100% amino acid sequence identity with that of S. aureus Z172 (Uniprot: A0A0E1AGX5). The S. aureus Z172 is also a ST239-SCCmec III hospital-associated MRSA, isolated in Taiwan, whose genome sequence had already been deposited in the GenBank (accession numbers: CP006838, CP006839, and CP006840). Whole genome alignment revealed a single inversion of approximately 0.4 Mbp between HC1335 and BMB9393 chromosomes, whose sequence is highly conserved between these genomes (fig. 1). Point mutation analysis revealed only three high quality single nucleotide polymorphisms (SNPs) in coding regions, all of them leading to non-conservative amino acid changes. The three predicted CDS affected by these substitutions showed putative functions of AraC family of transcriptional regulator (SAHC1335_00882; amino acid substitution Y456H in the hydrolase domain) and Carbamoyl-phosphate synthase large chain (SAHC1335_02130; amino acid substitution P289L in the phosphate transferase domain), in addition to a MOSC domain-containing protein (SAHC1335_00312; amino acid substitution Y25S at five amino acids from the sulfurase domain).
Fig. 1.—
Circular map of the chromosome of the S. aureus subsp. aureus isolates BMB9393 and HC1335. The innermost ring represents the BMB9393 chromosome used as reference and its coordinates. The second ring (in black) plots the G + C content of the reference, followed by its G + C skew (in purple/green). The following colored ring (dark pink) depicts BLASTN comparison between the chromosomes of BMB93933 and HC1335 isolates. The outer line (in black) represents the region of the predicted inversion between BMB9393 and HC1335 chromosomes.
Bacteriophage analyses detected five phage elements in the HC1335 genome, the bacteriophages ϕNM3-like, YMC/09-like and ϕMR25-like as intact prophages, the PT1028-like as a questionable prophage and an incomplete phage region probably related with the staphylococcal phage 96. Phage ϕNM3-like, is a β-hemolysin converting prophage, and carries the genes chp, scn and sak, encoding for the chemotaxis-inhibiting protein-CHIPS (de Haas et al. 2004), staphylococcal complement inhibitor-SCIN (Rooijakkers et al. 2005) and staphylokinase-SAK (Sako and Tsuchida 1983), respectively. These genes are part of innate immune evasion cluster (IEC) and provides S. aureus with a unique mechanism to adapt to and counteract the human host (Van Wamel et al. 2006). However, the sea gene encoding the staphylococcal enterotoxin A—frequently carried by this phage—was absent in the HC1335 genome. None confirmed CRISPR repeat was found in the chromosome of this isolate.
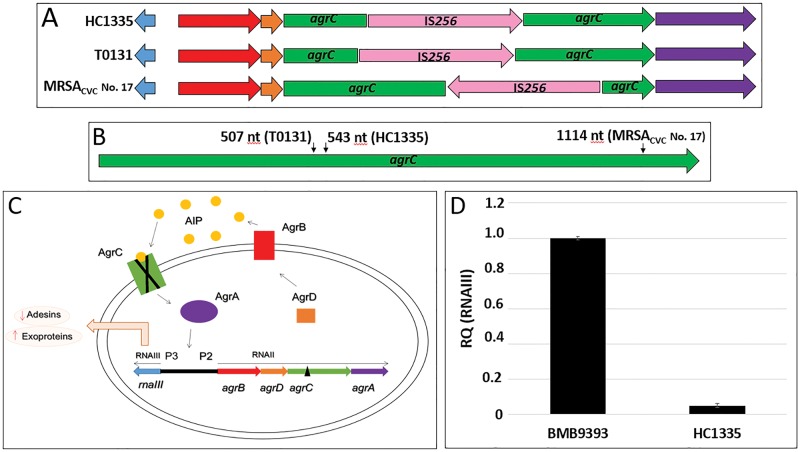
The search for possible dysfunction in virulence regulation caused by transposase insertions revealed that HC1335 isolate has a transposase element—derived from the insertional element (IS) named IS256—inserted in the agrC gene (fig. 2A) at nucleotide 543 (fig. 2B), which was absent in the BMB9393 genome. This insertion truncated the agrC with consequent inhibition of the gene, affecting the regulation of the whole agr locus (fig. 2C), as expected and confirmed by the experiments of RNAIII expression (fig. 2D). The IS256 element was initially characterized by Byrne et al. (1989) as part of the Tn4001. This element has 1324 bp in length and carries a single CDS (encoding a 390-amino acid transposase) preceded by a potential RBS (ribosome biding site) and −35 and −10 promoter sequences. Inverted repeat (IR) sequences were found upstream and downstream the transposase gene (tnp) (Byrne et al. 1989). The IS256-derived element found disrupting agrC in HC1335 has the same structure of the IS256 described by Byrne et al. (1989) but lacks part of the upstream and all the downstream IRs.
Fig. 2.—
Disruption of the agrC gene in MRSA isolates by IS256 insertions. (A). Schematic representation of the agr locus of the MRSA isolates HC1335 (this work), T0131 (Li et al. 2011) and MRSAcvc No 17 (Cafiso et al. 2007). The agrBDCA, rnaIII, and IS256 tnp coding sequences (CDSs) are represented by red, orange, green, purple, blue, and pink arrows, respectively). These isolates have an IS256 sequence inserted in the agrC CDS at different insertion sites, and in opposite direction in the case of the isolate MRSAcvc No 17. (B) Green arrow represents the agrC CDS and small black arrows indicate the positions of the insertion of the IS256 element in the isolates HC1335, T0131, and MRSAcvc No 17. (C) Schematic representation of the Agr system in S. aureus based on Novick et al. (2000). Activation of the P2 promoter leads to the transcription of the RNAII region that includes the genes agrBDCA. agrD encodes the AgrD protein (orange square) that is transported and processed by the AgrB (red rectangle) in small auto-inducer peptides, the AIP (yellow circles). Outside the cell, AIP binds to the histidine kinase receptor AgrC (green rectangle), which phosphorylates the response regulator AgrA (purple ellipse). The phosphorylated AgrA (P-AgrA) binds to both P2 and P3 promoters activating the transcription of the RNAIII—the main regulatory molecule of Agr system. Note that an insertion of a transposase element (black triangle) in the agrC (green rectangle) will produce a non-functional (X marked) AgrC protein, blocking the whole agr auto-induced circuit. (D) Expression of agr-RNAIII transcripts in the isolates BMB9393 (agr functional) and HC1335 (agr dysfunctional). The RNA preparation of the isolate BMB9393 was used as the calibrator sample for normalizing the experiment. RQ: relative quantification.
It is remarkable that we also found an IS256 element truncating the agrC gene (fig. 2A) in the genome of the ST239-SCCmecIII isolate T0131 (GenBank accession number: CP002643.1) recovered in 2016 from an 87-year-old patient in China (Li et al. 2011). In the case of the Chinese isolate, the complete IS256 element was inserted at nucleotide 507, i.e. 36nt upstream the insertion site observed in the HC1335 genome (fig. 2B). Notably, an IS256 element was also found inserted at nucleotide position 1114 of the agrC gene of another MRSA strain (Cafiso et al. 2007); however, in this case, the tnp was located in the agrC antisense strand (fig. 2B). Additionally, other authors have reported the insertion of IS256 elements in the agrC of staphylococci presenting distinct genetic backgrounds including non-ST239 S. aureus and Staphylococcus epidermidis isolates. However, neither agr locus sequence nor the IS256 position in the agrC were informed in those studies (Vuong et al. 2004; Shopsin et al. 2010; McEvoy et al. 2013). Taken together, these data support the idea of a convergent evolution toward agrC inactivation because the same genetic event—agr inactivation via agrC IS256-truncation—have paralleled occurred at different integration sites in distinct S. aureus lineages and even in different staphylococcal species (S. aureus and S. epidermidis), which, although have evolved in the same direction—leading to the inactivation of the whole Agr system—have most likely acquired the same characteristic independently. Therefore, this mechanism of agrC inactivation was likely driven by staphylococcal pathoadaptation, because Agr system is a global regulator of a plethora of virulence genes.
The presence of multiple copies of the IS256 is a common feature in MRSA genomes (McEvoy et al. 2013; Benson et al. 2014). In addition to the IS256-derived element inserted in the agrC, 20 intact copies of the IS256 element were detected in the chromosome of HC1335. In fact, Kwon et al. (2008) have positively correlated the prevalence IS256 in S. aureus genome with high-level multiresistance and biofilm capability. In addition, studies have detected the emergence of intermediate vancomycin resistance—probably driven by the selective pressure caused by the vancomycin treatment—due to IS256 insertions in the untranslated region (5′ UTR) of the autolysin regulator walKR, which reduced the regulator activity by 50% (Howden et al. 2010; McEvoy et al. 2013).
Benson et al. (2014) have reported an IS256 element in the genome of another MRSA clone, the so-called USA500, close to the region of the promoter sequence of rot regulator gene encoding the repressor of toxin (Rot). Those authors concluded that the insertion of the IS256 element in rot promoter region increases the expression of cytotoxins, including α-hemolysin (hla), with consequent increased virulence in an animal model (Benson et al. 2014).
Accordingly, the inactivation of rot and agr by the IS256 suggests that the insertion of this element in master gene regulators of successful MRSA clones (ST239 and USA500) may represent a customary mechanism of bacterial adaptability to different environmental and epidemiological scenario. Thus, taken together these comparative genomic studies have revealed a new role for the IS256 that by causing small change in MRSA genomes leads to an immediate reprogramming of the bacterial virulence repertory, with predictable consequences for bacterial colonization, spread, host-interaction and virulence potential.
Despite the importance of Agr in the pathogenesis of staphylococcal diseases, isolates lacking detectable Agr activity have also been collected from cases of S. aureus bacteremia (Fowler et al. 2004; Traber et al. 2008; Shopsin et al. 2010; Butterfield et al. 2011; Ferreira et al. 2012). Indeed, agr-defective S. aureus are more likely to cause persistent infections than the agr-functional isolates, resulting in increased rate of secondary infections and mortality (Fowler et al. 2004; Traber et al. 2008; Valour et al. 2015). Finally, the increased fitness observed among agr-dysfunctional isolates (Paulander et al. 2012) may represent a compensatory mechanism for the biological cost resulted from the acquisition of high number of multiresistant traits by ST239 hospital isolates. Actually, agr-dysfunction was also described in high percentage among ST239 hospital isolates from the State of Victoria, Australia (Baines et al. 2015).
Material and Methods
Genomic DNA was prepared from 50 mL trypticase soy broth (TSB) culture—37 °C for 18 h under vigorous shaking (250 RPM)—using phenol extraction and ethanol precipitation (Sambrook et al. 1989). The concentration and purity of the DNA preparation were assessed using a Qubit® 2.0 fluorometer (Invitrogen; Eugene, Oregon, USA) and 5 µg genomic DNA was used to prepare the library. Whole-genome sequencing was performed using a 454 GS FLX titanium (3-kb paired-end library) approach (Roche Diagnostics Corporation, Indianopolis, IN, USA). The assembly, based on 488,230 reads corresponding to 82,521,347 bp (27-fold coverage), was performed using Newbler v 2.6 (Roche) (Margulies et al. 2005) and Celera genome assembly v 6.1 from JCV Institute (Myers et al. 2000). High quality adapter-free reads were used for genome assembly. Optical duplicates were removed using the wrapper extract replicates that employs CD-HIT. After de-duplication, 315.034 reads were assembled using Newbler v 2.6 (Roche). Assembled reads have mean size of 169pb, and 95% of the bases on the assembled genome have Phred quality >40, calculated by Newbler. Gaps within scaffolds, resulting from repetitive sequences, were resolved by in silico gap filling. In order to perform this procedure, gaps intra- and inter-scaffolds were filled by individual assemblies of the reads present in both frontiers of the gap. This was accomplished by selecting reads that formed the end of contigs adjacent to each gap stretches. Those reads were assembled separately using Newbler v 2.6 (Roche). Contigous sequences generated by this approach that were able to complete the gap and anchor on the two adjacent contigs were added to the sequence, and thus closing the gap. This approach generated a gap- and error-free chromosome with only six ambiguous bases on the same homopolimeric-T stretch.
For gene prediction, it was applied the prediction programs Glimmer GeneMark and Prodigal (http://prodigal.ornl.gov/; last accessed July 1, 2016) including a correction routine for start codon from the multiple alignment of “n” similar proteins identified by BLASP against NR database (http://www.ncbi.nlm.nih.gov/protein; last accessed July 1, 2016). CDSs identified after this process were annotated (Gene, product, EC number etc.) using UniProt, Kegg and NR databases. In this process, it was only considered the CDSs over 70 aa with almost one BLASTP hit against any of the three databases mentioned above. Next, a comparative analysis was done for gene content between HC1335 and BMB9393 using BLASTP (1e−05 E value), using all CDSs of the genome of HC1335 against all CDSs of the genome of BMB9393. Only genes with no hit were considered, in order to find exclusive genes in HC1335. Genome annotation was performed using SABIA pipeline (Almeida et al. 2004). Genes encoding rRNA and tRNA were identified with RNAmmer (Lagesen et al. 2007) and tRNAscan (Schattner et al. 2005), respectively. Genome map was drawn using BRIG (Alikhan et al. 2011). Bacteriophage analysis was performed using Phage Search Tool (PHAST) (Zhou et al. 2011). Comparative analysis was conducted using SABIA comparative tool (Almeida et al. 2004) and whole-genome alignment using Mauve (Darling et al. 2010). SNP calling was performed using the BMB9393 genome as reference. Briefly, the deep sequencing libraries files were quality checked with FASTQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; last accessed July 29, 2016) and further trimmed with Fastx_toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html; last accessed July 29, 2016). The trimmed reads from HC1335 were mapped against the BMB9393 genome using the Bowtie 2 mapper with one mismatch per seed region (20 nt in length). The resulting mapping files were treated with SAMtools program (http://samtools.sourceforge.net/; last accessed July 29, 2016), only mapping reads with map quality (mapQ) above 30 were kept. The Picard mark duplicates tool (http://broadinstitute.github.io/picard/; last accessed July 29, 2016) was used in order to flag putative sequencing artifact, such as optical duplicates. The Genome Analysis Toolkit—GATK (https://software.broadinstitute.org/gatk/; last accessed July 29, 2016) was used to call the variants using default parameters. The SNPs were annotated with the snpEFF tool (http://snpeff.sourceforge.net/; last accessed July 29, 2016) and custom Python scripts. Only SNPs with coverage bigger than 10 reads were considered in this analysis.
For RNA preparations, bacterial cells grown in brain-heart infusion (BHI) broth (18 h; 37 °C; 250 rpm) were collected in the stationary phase. The total RNA was prepared using the RNeasy Mini kit (Qiagen, Maryland, IN, USA) and quantified by Nanodrop Lite (Thermo Fisher Scientific, Wilmington, DE, USA). The RNA quality was analyzed using RNA gel electrophoresis. Real time quantitative reverse transcription polymerase chain reaction (real time qRT-PCR) was performed using the Power SYBRW Green RNA-to-CT TM 1-Step Kit (Applied Biosystems, Foster City, CA, USA) as recommended, and the amount of transcripts were estimated by the Ct comparative method. The primers used for agr-RNAIII and 16S rRNA (endogenous control) were described previously (Ferreira et al. 2012). The run was performed in a Step OneTM Real Time PCR System (Applied Biosystems) and analyzed using the Step One Software 2.2 (Applied Biosystems).
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported in part by the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ grant no.: E26/102.901/2011, E26/110.625/2011 and E26/111.663/2013); and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grant no. 472034/2012-0). Botelho AMN was the recipient of a fellowship from Fundação CAPES/FAPERJ.
Literature Cited
- Alikhan N-F, Petty NK, Ben Zakour NL, Beatson S. A. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida LGP, et al. 2004. A System for Automated Bacterial (genome) Integrated Annotation - SABIA. Bioinformatics 20:2832–2833. [DOI] [PubMed] [Google Scholar]
- Baines SL, et al. 2015. Convergent adaptation in the dominant global hospital clone ST239 of methicillin-resistant Staphylococcus aureus. 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M., et al. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol. Microbiol. 93:664–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield JM, et al. 2011. Predictors of agr dysfunction in methicillin-resistant Staphylococcus aureus (MRSA) isolates among patients with MRSA bloodstream infections. Antimicrob. Agents Chemother. 55:5433–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Rouch DA, Skurray RA. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361–367. [DOI] [PubMed] [Google Scholar]
- Cafiso V, et al. 2007. agr-Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 51:220–227. [DOI] [PubMed] [Google Scholar]
- Campos GB, et al. 2012. Isolation, molecular characteristics and disinfection of methicillin-resistant Staphylococcus aureus from ICU units in Brazil. New Microbiol. 35:183–190. [PubMed] [Google Scholar]
- Costa MO, et al. 2013. Complete genome sequence of a variant of the methicillin-resistant Staphylococcus aureus ST239 lineage, strain BMB9393, displaying superior ability to accumulate ICA-independent biofilm. Genome Announc 1:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira FA, et al. 2012. Comparison of in vitro and in vivo systems to study ica-independent Staphylococcus aureus biofilms. J. Microbiol. Methods 88:393–398. [DOI] [PubMed] [Google Scholar]
- Figueiredo AMS, Ferreira FA. 2014. The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem. Inst. Oswaldo Cruz. 109:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VG, et al. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190:1140–1149. [DOI] [PubMed] [Google Scholar]
- de Haas CJC, et al. 2004. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden BP, et al. 2010. Complete genome sequence of Staphylococcus aureus strain JKD6008, an ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate-level vancomycin resistance. J. Bacteriol. 192:5848–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon AS, et al. 2008. Higher biofilm formation in multidrug-resistant clinical isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 32:68–72. [DOI] [PubMed] [Google Scholar]
- Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le KY, Otto M. 2015. Quorum-sensing regulation in staphylococci—an overview. Front. Microbiol. 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. 2012. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. 2011. Complete genome sequence of Staphylococcus aureus T0131, an ST239-MRSA-SCCmec type III clone isolated in China. J. Bacteriol. 193:3411–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins A, Moraes Riboli DF, Cataneli Pereira V, de Lourdes Ribeiro de Souza da Cunha M. 2014. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from a Brazilian university hospital. Braz. J. Infect. Dis. 1:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy CRE, et al. 2013. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of walKR expression. Antimicrob. Agents Chemother. 57:3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, et al. 2000. A whole-genome assembly of Drosophila. Science 287:2196–2204. [DOI] [PubMed] [Google Scholar]
- Novick R, Ross H, Figueiredo A, Abramochkin G. 2000. Activation and Inhibition of staphylococcal agr system. Science (New York, N.Y.). 287:391. [Google Scholar]
- Paulander W, et al. 2012. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. MBio 3:e00459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck SY, et al. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijakkers SHM, et al. 2005. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6:920–927. [DOI] [PubMed] [Google Scholar]
- Sako T, Tsuchida N. 1983. Nucleotide sequence of the staphylokinase gene from Staphylococcus aureus. Nucleic Acids Res. 11:7679–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritch E, Maniatis T. 1989. Commonly used techniques in molecular cloning In: Sambrook J, Fritch E, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; pp. E3–E4. [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Eaton C, Wasserman GA, et al. 2010. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202:1593–1599. [DOI] [PubMed] [Google Scholar]
- Teixeira LA, et al. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber KE, et al. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valour F, et al. 2015. Delta-toxin production deficiency in Staphylococcus aureus: a diagnostic marker of bone and joint infection chronicity linked with osteoblast invasion and biofilm formation. Clin. Microbiol. Infect. 21:568.e1–568.e11. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190:1498–1505. [DOI] [PubMed] [Google Scholar]
- Van Wamel WJB, Rooijakkers SHM, Ruyken M, Van Kessel KPM, Van Strijp JAG. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39: W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.