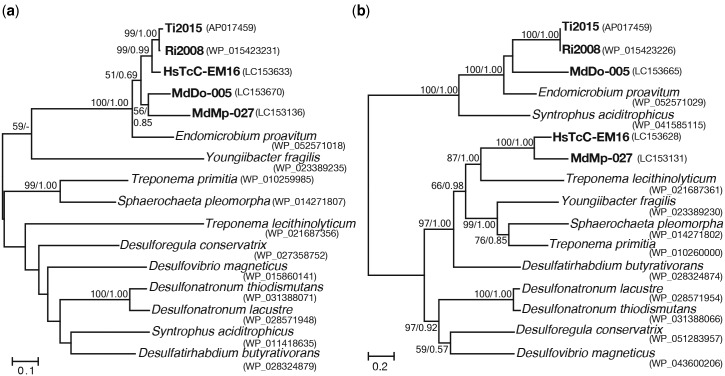
Fig. 4.—
Phylogenetic positions of the Cas1 and Cas3 proteins in the type I-C CRIPSR/Cas systems of the phylum Elusimicrobia. Maximum-likelihood trees were constructed using the LG + Γ and LG + Γ + I amino acid substitution models, for Cas1 and Cas3, respectively. Bootstrap confidence values (left) and posterior probabilities in Bayesian statistics (right) are shown for the internal branches. Only values >50% are shown. Sequences obtained in this study are shown in bold. (a) Cas1 protein. 341 unambiguously aligned sites were used. (b) Cas3 protein. 541 unambiguously aligned sites were used. The pseudogene of cas3 in Endomicrobium proavitum was included by adjusting codon frames.