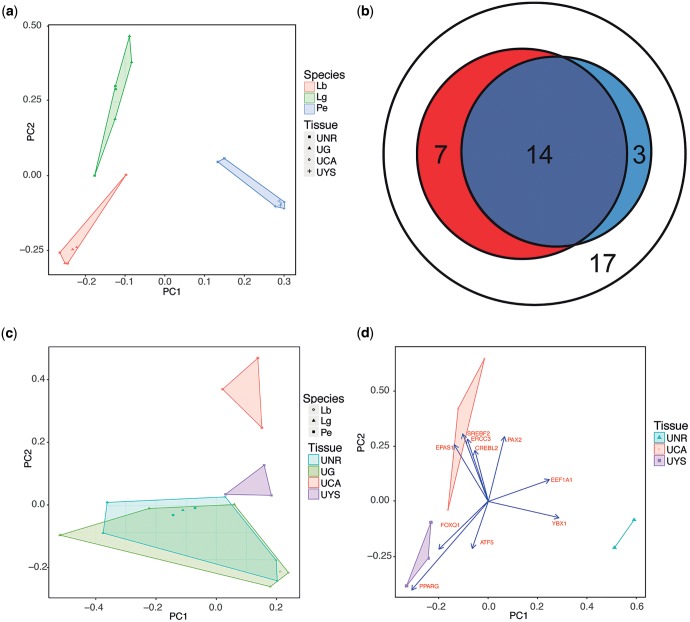
Fig. 2.—
Comparisons of placental gene expression between different species and placental region. (a) PCA of square root TPM for all shared genes in the uterine tissue of the oviparous skinks Le. bougainvillii (Lb) and La. guichenoti (Lg), and the viviparous skink P. entrecasteauxii. Uterine tissue is from either non-reproductive (UNR) or gravid (UG) females, or from the chorioallantoic (UCA) or yolk sac (UYS) placenta of late pregnant females. Samples form discrete clusters based on the species of origin. (b) Amino acid transporter gene use in the placentae of reptiles and mammals. There is an overlap in the use of orthologous amino acid transport protein genes by human trophoblast (red) and grass skink uterine tissue (blue) during pregnancy. 41 orthologous amino acid transport proteins are shared by both lineages, 21 are utilized in human placenta, while 17 are used in grass skinks. About 17 genes are not utilized in the placental tissues of either species (white). Lineages show a significantly non-random use of genes (P = 0.003, hypergeometric test). (c) PCA of square root TPM for amino acid transporter gene expression in the uterine tissue of oviparous and viviparous skinks. In P. entrecasteauxii, uterine tissue from each placental tissue clusters with each other based on amino acid transporter expression, and there is an overlap in the expression profiles of gravid oviparous uterine tissue and non-pregnant uterine samples from all species. (d) PCA of square root TPM for transcription factors expressed in the uterine tissue of P. entrecasteauxii. Three discrete clusters form one for each placental region, and one for non-pregnant tissue. The data are overlaid with the loadings for the top 10 genes that contribute to the distance between samples in these principle components.