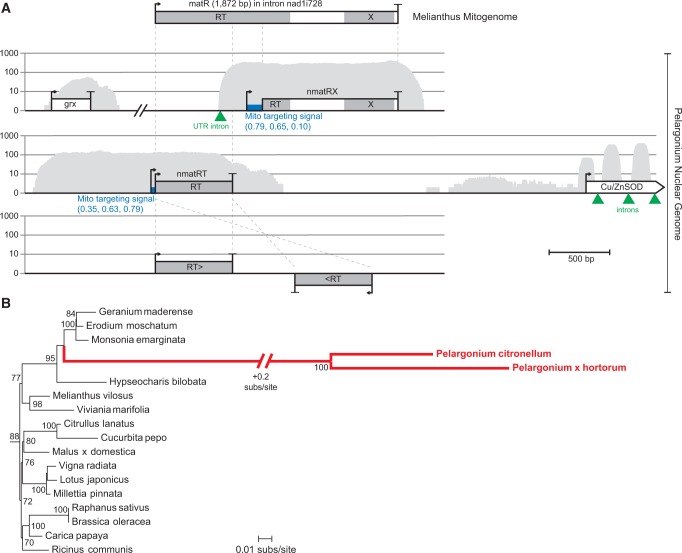
Fig. 4.—
Nuclear-encoded nmatR genes in Pelargonium. (A) The mitochondrial matR gene in the nad1 intron nad1i728 in M. villosus is shown at top, and homologous matR sequences in contigs from the Pelargonium nuclear genome are shown below. Start and stop codons are indicated by the arrow and bar, respectively. Grey shading shows conserved reverse transcriptase (RT) and maturase (X) domains. The thick blue bar following the first potential start codon of the nmatR sequences defines the location of the acquired mitochondrial targeting sequence; targeting prediction scores for MitoProt, TargetP, and Predotar are shown in blue in parentheses below the target sequence. The gene expression pattern of the nuclear contigs is mapped in grey. The location of nuclear spliceosomal introns are marked with a green triangle. All gene arrangements were drawn to scale. (B) Excerpt of a phylogenetic tree of the mitochondrial matR gene of angiosperms (thin black branches) and the nuclear nmatR paralogs of Pelargonium (thick red branches). All bootstrap values >70% are shown at respective nodes in the tree. The full phylogenetic tree is presented in supplementary figure S1, Supplementary Material online.