Abstract
Human skin has evolved rapidly, leaving evolutionary signatures in the genome. The filaggrin (FLG) gene is widely studied for its skin-barrier function in humans. The extensive genetic variation in this gene, especially common loss-of-function (LoF) mutations, has been established as primary risk factors for atopic dermatitis. To investigate the evolution of this gene, we analyzed 2,504 human genomes and genotyped the copy number variation of filaggrin repeats within FLG in 126 individuals from diverse ancestral backgrounds. We were unable to replicate a recent study claiming that LoF of FLG is adaptive in northern latitudes with lower ultraviolet light exposure. Instead, we present multiple lines of evidence suggesting that FLG genetic variation, including LoF variants, have little or no effect on fitness in modern humans. Haplotype-level scrutinization of the locus revealed signatures of a recent selective sweep in Asia, which increased the allele frequency of a haplotype group (Huxian haplogroup) in Asian populations. Functionally, we found that the Huxian haplogroup carries dozens of functional variants in FLG and hornerin (HRNR) genes, including those that are associated with atopic dermatitis susceptibility, HRNR expression levels and microbiome diversity on the skin. Our results suggest that the target of the adaptive sweep is HRNR gene function, and the functional FLG variants that involve susceptibility to atopic dermatitis, seem to hitchhike the selective sweep on HRNR. Our study presents a novel case of a locus that harbors clinically relevant common genetic variation with complex evolutionary trajectories.
Keywords: copy number variation, structural variants, positive selection, skin evolution, tandem repeats
Introduction
The skin acts as a first barrier against pathogens, biotic and abiotic agents (Proksch et al. 2008). Skin also helps regulate internal body temperature (Kraning 1991) and regulates ultraviolet (UV) radiation penetration (Jablonski and Chaplin 2000). Overall, skin interacts with the environment at multiple levels, potentially exposing it to various adaptive forces (Jablonski 2012).
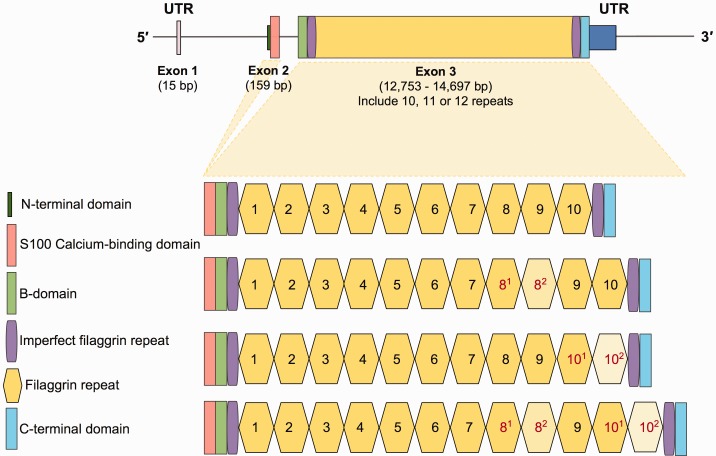
The human filaggrin (FLG) gene (NCBI RefSeq Accession NM_002016.1) is one of the best studied members of Epidermal Differentiation Complex (Mischke et al. 1996). Its organization is similar to the other members of evolutionarily related S100 fused-type proteins (SFTP) (Kypriotou et al. 2012) (fig. 1A). Briefly, FLG is organized into three exons and two introns (Gan et al. 1990). Exon 1 (15 bp) is noncoding and the small exon 2 (159 bp) encodes for 46 amino acids that encompass the translation start site and the calcium-binding domain of the protein. Exon 3, one of the largest in the human genome (∼12.7 kb), encodes 10 nearly identical 972-bp-long tandem filaggrin repeats. In addition, exon 3 encodes for two imperfect repeats flanking the complete repeats, a B-domain and a unique tail sequence (Presland et al. 1992).
Fig. 1.—
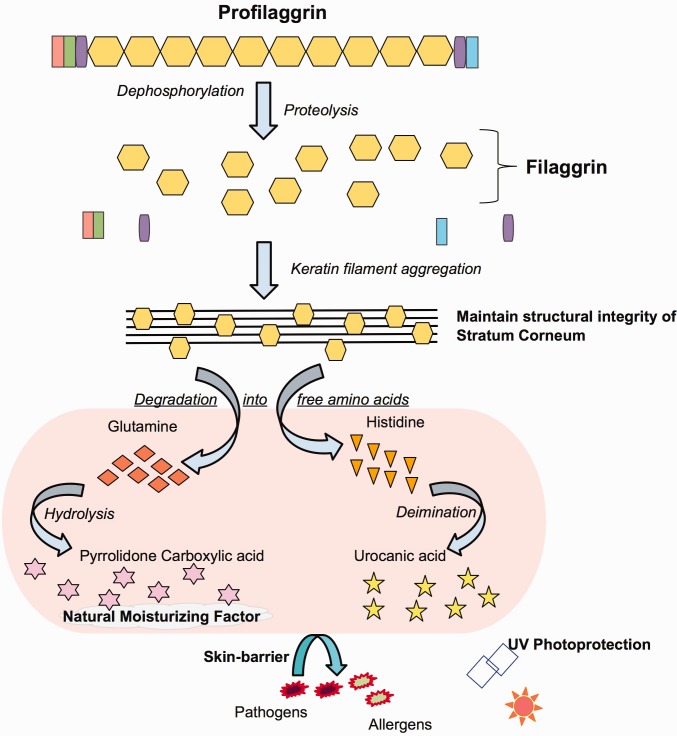
(B) Profilaggrin processing and function. This figure shows different roles of structural units of the FLG as color-coded in figure 1A in skin formation and function.
Fig. 1.—
(A) Gene structure and organization. The top figure shows the organization of FLG. The thin line indicates introns and the thicker lines indicate exonic sequences. The 5′- and 3′-UTRs, and the two coding exons are labeled and with the sizes in base pairs based on reference genome is indicated. The organizational/functional units of the gene are color-coded as shown on the lower left of the figure. At the lower part of the figure, genetic variation involving subexonic filaggrin repeats is shown.
FLG encodes for a protein with a complicated life-cycle and multiple functions primarily in skin (Brown and McLean 2012) (fig. 1B). The initial protein product, profilaggrin, is post-translationally cleaved into individual filaggrin peptides that were shown to have a dual role in skin. On the one hand, the individual filaggrin peptides bind to the keratin filaments and aggregate within the cytoskeleton of the keratinocytes condensing into tight bundles (Steinert and Marekov 1995; Simon et al. 1996). This step seems to contribute to the maintenance of cohesion and constancy among corneocytes, which form the skin-barrier, thereby preventing transepidermal water loss and shielding against the invasion of external factors such as pathogenic microbes and allergenic molecules (Candi et al. 2005; Angelova-Fischer et al. 2011). In simpler terms, FLG helps upkeep the structural integrity of the skin in mammals (Dale et al. 1978). Furthermore, the individual filaggrin peptides is further degraded into free amino acids, which in turn further broken down into urocanic acid and pyrrolidone carboxylic acid (Scott et al. 1982). These smaller molecules were shown to guard skin from UV radiation (Mildner et al. 2010; Barresi et al. 2011) and contribute to the natural moisturizing factor of the skin (Rawlings and Harding 2004).
Unexpectedly, there is a remarkable amount of functional genetic variation affecting FLG. Most dramatically, several common and population-specific loss-of-function (LoF) variants were reported in European (Palmer et al. 2006), Asian populations (Hsu et al. 2009; Chen et al. 2011). Very few LoF variants have been identified in people with African descent so far (studies were conducted only in African Americans) (Winge et al. 2011; Margolis, Gupta, Apter, Hoffstad, et al. 2014). The FLG LoF variants lead to impaired skin-barrier function. This impairment, in turn, predisposes the individual to dry, itchy, red skin and transepidermal water loss, which are major susceptibility factors for several complex skin disorders, including ichthyosis vulgaris (Smith et al. 2006; Sandilands et al. 2007; Gruber et al. 2011) and eczema/atopic dermatitis (Sandilands et al. 2007; Gao et al. 2009). In addition to LoF variants, filaggrin repeats were shown to be copy number variable ranging from 10 to 12 copies among human populations. The copy number of these repeats was negatively associated with atopic dermatitis susceptibility (Brown et al. 2012). However, a comprehensive documentation of the global distribution of FLG genetic variation free of ascertainment bias has yet to be compiled.
Despite its etiological relevance to skin disease, the evolution of FLG genetic variation remains unexplored. FLG resides in a chromosomal cluster of skin-related genes, which is a hotspot for evolutionary innovation involving skin adaptation (Strasser et al. 2014). For instance, one of these genes, RPTN, which encodes an epidermal matrix protein and is similar to FLG in sequence and in exonic-repeat structure, carry multiple fixed amino acid changes between humans and Neandertals (Green et al. 2010). Similarly, FLG2, the paralogous neighboring gene to FLG harbor very frequent loss-of-function variation, which was associated with atopic dermatitis (Margolis, Gupta, Apter, Ganguly, et al. 2014). The same gene was shown to have multiple duplicates in rhesus macaques as compared with great apes (Gokcumen et al. 2013). Upstream of FLG, variation affecting late cornified envelope genes, were associated strongly with susceptibility to psoriasis (de Cid et al. 2009). From an evolutionary point-of-view, one of these psoriasis-associated variants, a large deletion overlapping with LCE3B and LCE3C genes, was shown to predate the Human–Neandertal divergence (Lin et al. 2015) and argued to be evolving under balancing selection (Pajic et al. Under review). Despite this interesting company, very few studies attempted to explore the recent evolution of FLG in humans. In this study, we resolved the haplotype structure that defines FLG genetic variation and investigated the evolution of this gene using population genetic analyses of thousands of genomes.
Results
The Frequency of FLG Loss-of-Function Variants Does Not Correlate with Latitude
Loss-of-function (LoF) variants affecting FLG are common. They have been extensively studied in Eurasian populations due to their strong association with atopic dermatitis (Palmer et al. 2006) and ichthyosis vulgaris (Smith et al. 2006), as well as other skin diseases (reviewed in Winge and Bradley 2014). The evolutionary reasons as to why these disease-causing LoF mutations are common in humans remains unknown. A recent study put forward a hypothesis that claimed that loss-of-function of FLG is adaptive in environments with low ultraviolet light (Thyssen et al. 2014). This study is based on the above-described functional work showing that side products of FLG can act as ultraviolet (UV) light absorbers (fig. 1B). Adequate penetration of skin by UV light is important for vitamin D metabolism (Holick et al. 2007). As such, the authors hypothesized that LoF variants of FLG may be adaptive where UV light is not abundant. They then tested this hypothesis by conducting a meta-analysis of existing datasets on allele frequency of LoF mutations across the world. Their results indeed showed that the allele frequency of LoF mutations in FLG increases with latitude. This observation supports the notion that FLG LoF variants may be adaptively selected in northern latitudes, increasing their allele frequency especially in northern European populations.
We were intrigued by this hypothesis, but concerned with the fact that the datasets used by Thyssen et al. (2014) relied mostly on meta-analysis of known LoF variants, rather than unbiased variant-calling. Therefore, there is a possibility that their analysis suffers from ascertainment bias (Clark et al. 2005; Rosenberg et al. 2010), especially when they involve African populations (Lachance and Tishkoff 2013). To replicate the results of Thyssen et al. (2014), this time controlling for ascertainment bias, we used recently available 1000 Genomes Phase 3 dataset (1-kGP dataset, goo.gl/bCu3oC), which involve resequencing data from 2,504 samples across 26 populations (1000 Genomes Project Consortium et al. 2015). Unlike the previous attempts to characterize the genetic variation in FLG, 1-kGP dataset allows global discovery of both common (>5%) and intermediate (>1%) allele frequency variants without ascertainment bias across multiple populations. Using this dataset, we found 51 nonsense single nucleotide variants, including all of the common (>5% allele frequency) variants (13) that were observed by Thyssen et al. (2014) (fig. 2 and supplementary table S1, Supplementary Material online).
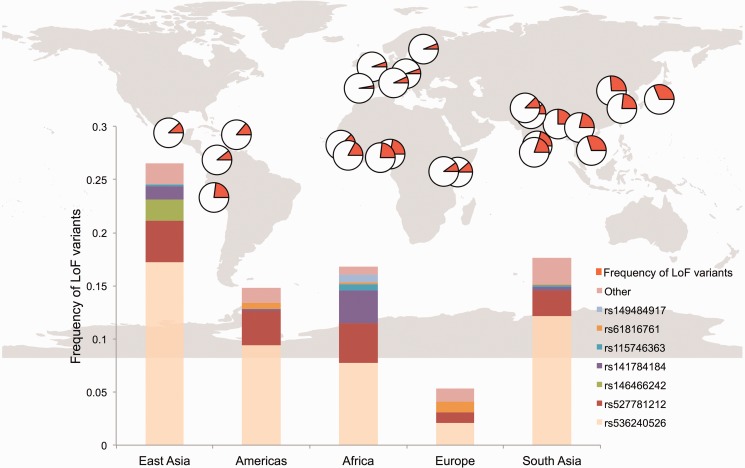
Fig. 2.—
Allele frequency distribution of FLG loss-of-function SNPs. The map on top shows the distribution of loss-of-function alleles across the world. The orange portion of the pies corresponds to cumulative allele frequency of loss-of-function alleles detected in the 1000 Genomes Phase 3 dataset. The bottom graph highlights the most common loss-of-function variants in different continental populations.
Based on this new dataset, we found no significant positive association between cumulative allele frequency of LoF variants and latitude (R2 = 0.1075; supplementary fig. S1A, Supplementary Material online). To ensure that our pipeline does not have a bias for generating false-negative results, we repeated our analysis with a nonsynonymous variant, rs16891982, which has known affect the level of melanin production and is strongly associated with UV protection (Stokowski et al. 2007). Indeed, we observed the expected correlation between the frequency of rs16891982 and latitude using 1000 Genomes Phase 3 data (R2 = 0.5368, supplementary fig. S1B, Supplementary Material online). We also replicated the lack of correlation between FLG LoF variants and latitude using the 1000 Genomes Phase 1 dataset, which used a different variant calling pipeline than the Phase 3 dataset did (R2 = 0.03726, supplementary fig. S1C, Supplementary Material online). In parallel, we also calculated the correlation of individual common LoF variants with latitude and found no correlation higher than R2 = 0.1. To ensure that our results do not reflect discovery errors in 1 kGP, we manually checked individual samples for misalignments in Integrated Genome Browser (http://bioviz.org/igb/index.html) and found no apparent errors (supplementary fig. S2, Supplementary Material online).
To further confirm our results, we verified that the allele frequencies of most of the LoF variants in the 1-kGP dataset are highly concordant with those in ExAC database (http://exac.broadinstitute.org/), which compiled exome sequencing data from tens of thousands of individuals (supplementary table S1, Supplementary Material online). We found that majority of the common (>0.1 allele frequency) LoF variants are consistently documented in both databases. We found 2 LoF variants (rs527781212, rs141784184) that are found in 1000 Genomes in slightly >0.01 allele frequency, but essentially absent in ExAC database. Thus, rs527781212 and rs141784184 may be false negatives in 1-kGP dataset. We found one LoF variant that is found in considerably different allele frequencies in 1-kGP and ExAC datasets in Asian populations (∼0.004 and 0.029 allele frequency, respectively). Further scrutinization shows that this variant is found primarily in Han Chinese and very infrequently in other Asian population (supplementary table S1, Supplementary Material online). As such, population specificity, rather than accuracy of the datasets, explain the difference in allele frequency between datasets. For a comprehensive picture of the rare functional variation, it may be necessary to have broader population sampling especially for rapidly evolving loci such as FLG. Regardless, to ensure that our overall analysis is not biased because of the disparities in the allele frequencies of individual LoF variants, we calculated cumulative allele frequency of LoF variants in continental populations using only ExAC database (supplementary fig. S3, Supplementary Material online). We observed no apparent correlation with latitude concordant with our observations using 1-kGP data. In fact, the LoF variants are in higher frequency in Africa. Overall, our results are robust and we found no association between latitude and the frequency of FLG LoF variants. Further scrutinization, described below, reveals that the lack of replication primarily stems from multiple population-specific LoF variants that were not analyzed by Thyssen et al. (2014) (supplementary table S1, Supplementary Material online).
FLG LoF Variants Do Not Have Observable Fitness Effects
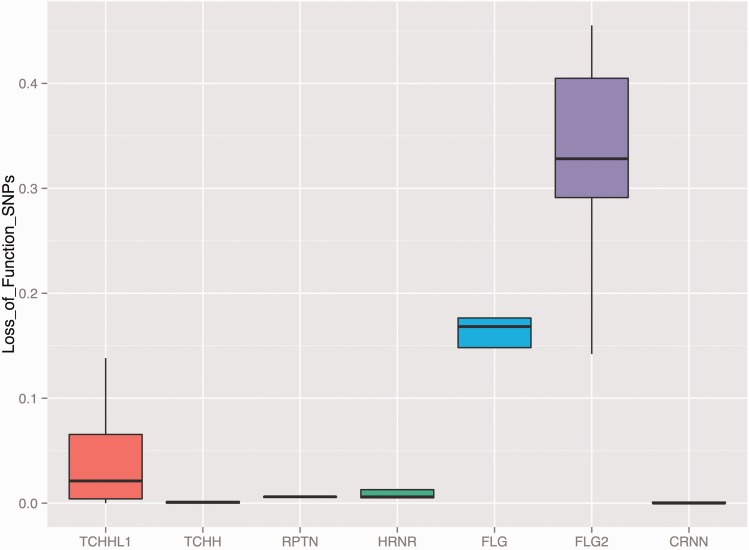
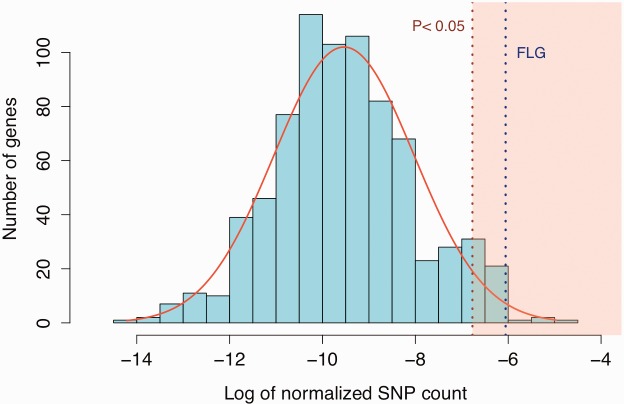
We investigated whether the number and frequency of FLG LoF variants are indeed unusual as compared with LoF variants in other genes. We first confirmed that the number of LoF in FLG is significantly higher than what is observed for other genes in the human genome (P = 0.04689, Wilcoxon rank sum test, fig. 3A). We also found that the frequency of FLG LoF variants are higher even when considered individually, when compared with other genes in the genome (P = 5.136e-16, Wilcoxon rank sum test, fig. 3C). The frequency of FLG LoF variants are indeed remarkably high when compared with other members of the SFTP gene family with the dramatic exception of FLG2 (fig. 3D). We considered the possibility that LoF mutations are clustered in the 3′-UTR of the FLG, and hence do not interfere with the main function of the gene, which is to provide filaggrin blocks to support epidermis structure. However, we observed no clustering of FLG LoF across the gene, even when allele frequencies are considered (fig. 3B). In fact, we also found loss-of-function variation in chimpanzees (1 out of 10 haplotypes, panTro3, chr1:130518034) in a previously published database (Gokcumen et al. 2013), suggesting the accumulation of LoF variations may have started before Human–Chimpanzee divergence.
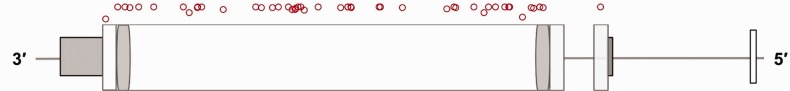
Fig. 3.—
(B) The distribution of individual loss-of-function variants in FLG gene. The red dots on top of the FLG gene model stand for the loss-of-function variants found in FLG.
Fig. 3.—
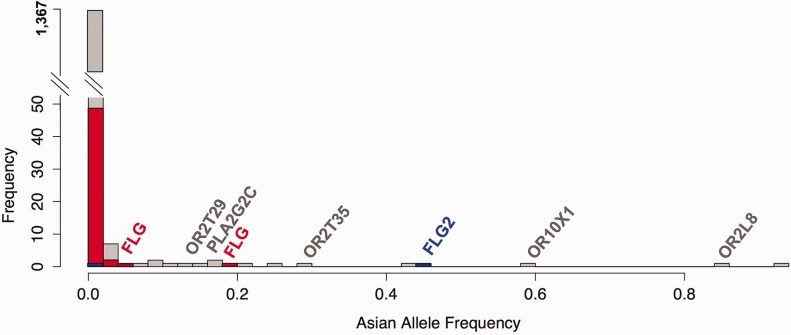
(C) The histogram for the frequencies of multiple FLG loss-of-function variants in the genome is significantly higher than chromosome-wide expectations. The x-axis of this histogram shows the allele frequency of observed loss-of-function variants in Asian population across chromosome 1. The y-axis shows the number of observations. The majority of loss-of-function variants are rare (<1%). However, four different FLG loss-of-function variants are individually have higher allele frequencies, and cumulatively the allele frequency of FLG loss-of-function variants represents one of the highest in the genome.
Fig. 3.—
(D) The frequency of SFTP gene family loss-of-function variants. This x-axis of the boxplot indicates the genes neighboring FLG. The y-axis indicates the cumulative allele frequency of loss-of-function variants affecting these genes in 1000 Genomes Phase 3 dataset populations. All the genes represented in this figure are members of the SFTP gene family and have similar gene organization with FLG, carrying large exonic repeats.
Fig. 3.—
(A) The number of FLG loss-of-function variants standout from the chromosome-wide expectations. The x-axis of this plot indicates the log number of loss-of-function variants observed in each gene (normalized by gene size), on chromosome 1 using the 1000 Genomes Phase 3 dataset. Note that genes with no loss-of-function variants reported were not plotted. The number of FLG loss-of-function variants is significantly higher than the chromosome-wide expectation.
The null hypothesis for such high frequency of loss-of-function variation is the reduction in the strength of purifying (negative) selection as exemplified by some of the olfactory receptor genes in primates (Rouquier et al. 2000) and protease genes in humans (Somel et al. 2013). In this scenario, genetic drift would be the major force that shapes the frequency and geographic distribution of FLG LoF variants in human populations. However, before reaching this conclusion, we considered adaptive forces favoring LoF variants in human populations. In its simplest form, as exemplified for the 577X null allele affecting ACTN3 gene (MacArthur et al. 2007), the positive selection would: (i) increase the allele frequency of a single LoF variant in a given population, increasing population differentiation, (ii) decrease the genetic variation in the locus, and (iii) leave a long-range linkage disequilibrium (LD) signature as exemplified elsewhere. To test these for the FLG locus, we compared the single nucleotide variants in FLG to the rest of chromosome 1 for any deviation from expected genome-wide distribution in linkage disequilibrium (iHS), population differentiation (FST) or nucleotide variation (pi) using available datasets (Pybus et al. 2014). We found no significant deviation from the expected genome-wide distribution for these tests for LoF variants (supplementary fig. S4A–C, Supplementary Material online). Overall, we were not able to reject the null hypothesis that FLG LoF variants have been evolving under relaxed (or completely absent) negative selection, and have no observable fitness effects despite their well-established role in skin disease. Therefore, our analyses suggest that the main evolutionary force that shape the extant distribution of FLG LoF variants is genetic drift.
Resolving Haplotype Structure around FLG Unravels a Haplogroup That Encompasses FLG and HRNR Genes and Shows Signatures of a Selective Sweep
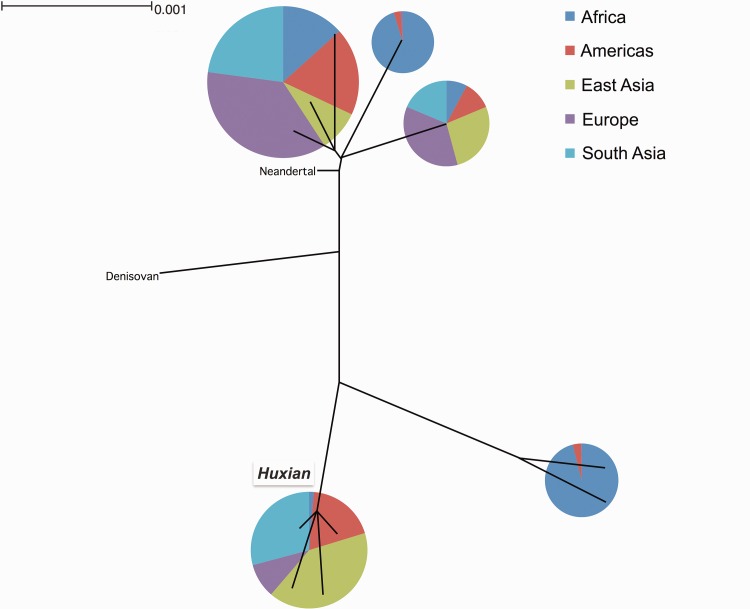
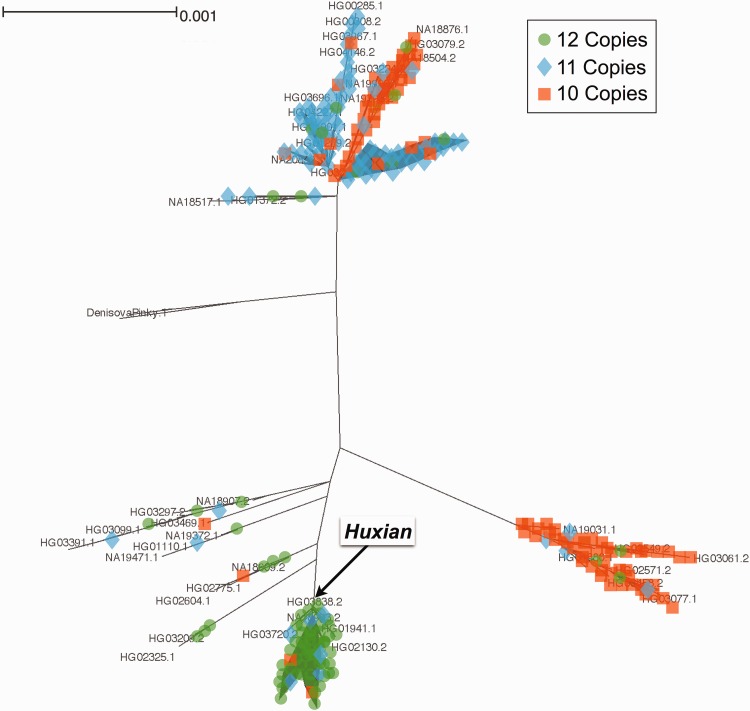
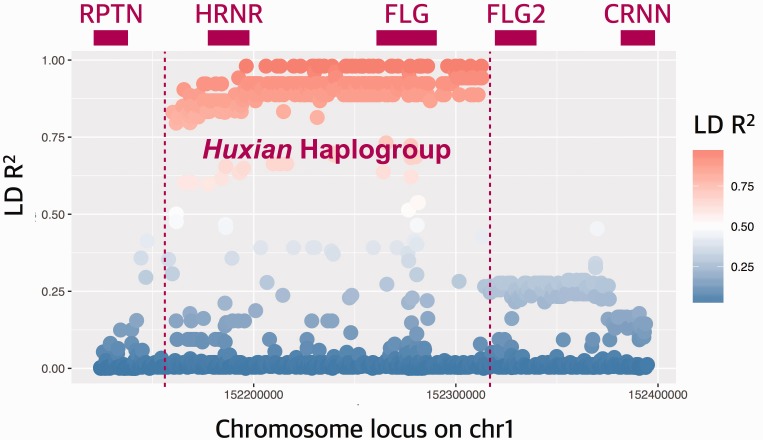
We were surprised to observe that some variants in FLG (other than LoF variants) have significantly higher iHS scores when compared together to the neutral portions of the genome. This trend is especially strong in the Chinese population (supplementary fig. S4A, Supplementary Material online). To further understand the underlying haplotype structure that led to high iHS values observed for the FLG locus, we searched the 1000 genomes dataset that are in near-perfect linkage disequilibrium with the haplotype in Asian populations using the single nucleotide variant rs77422831, which has the highest iHS value (2.75625) in the Chinese (CHB) population. We found 359 single nucleotide variants with strong (R2 > 0.8) linkage disequilibrium with rs77422831, constituting a haplotype block spanning not only FLG, but also the neighboring HRNR gene (fig. 4A). To contextualize this haplotype with the overall genetic variation in this locus, we constructed a maximum likelihood tree using the phased haplotypes from the 1000 Genomes Phase 3 dataset (fig. 4B). Note that we used 10-kb upstream of FLG, which captures the haplotypic variation affecting FLG locus, but avoids potential mapping biases due to segmental duplications and copy number variation within FLG. Our results revealed the haplotype group (named hereon as Huxian Haplogroup based on the Chinese folklore of the trickster fox spirit), which is defined by the high-iHS rs77422831 (the T allele tags Huxian haplotype) and is clearly separated from the rest of human haplotypes. The Huxian haplogroup exists most frequently in East Asia, but considerably less in Europe and Africa. In fact, this haplogroup represents the majority of haplotypes in CHB and also in other Asian populations (fig. 4C).
Fig. 4.—
(B) FLG Haplotype tree. This is the simplified maximum likelihood tree of all FLG haplotypes reported in the 1000 Genomes Phase 3 dataset. We constructed this tree using the sequence 10,000 bp immediately upstream of FLG. The pie-charts show the continental allele-frequency of each major haplotype group as color coded in the upper right side of the figure. The size of the pie-charts shows the relative number of haplotypes represented in each haplotype group. The Huxian haplogroup, as well as the Neandertal and Denisovan haplotypes were indicated.
Fig. 4.—
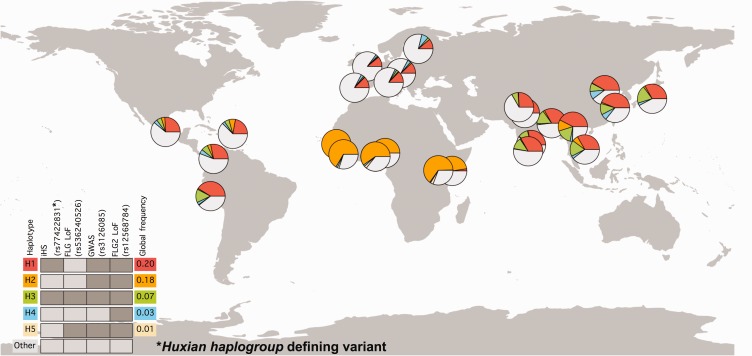
(C) Geographic distribution of haplotypes carrying functionally relevant variants. Distribution of most common combinations of putatively functional variants were shown across the world. The colors in the pie-charts represent the relative frequencies of major haplotypes. The individual SNPs that define different haplotypes were indicated on top of the legend placed bottom left of the map. The dark gray bars indicate the presence of the derived allele in that haplotype, whereas light-gray indicate that the haplotype carry the ancestral allele. The color-coded haplotypes indicated by H1–H5 corresponds to the colors in the pie-charts. Other indicate haplotypes not covered in this chart. The global allele frequencies of each haplotype were indicated in the right side of the legend. The H1 (red) and H3 (green) haplotypes correspond to the Huxian haplogroup. Note the major increase in allele frequency of the Huxian haplogroup in Eastern Asian and Southern Asian populations.
Fig. 4.—
(D) FLG maximum likelihood tree. We constructed a tree for the region 10,000 bp immediately upstream of FLG based on the 1000 Genomes Phase 3 dataset. It is the basis of the more simplified version represented in figure 4A. In addition, we genotyped 126 samples (252 haplotypes) for filaggrin copy number and manually imputed the haplotypes carrying these repeats. We indicated the imputed copy numbers onto this tree where we indicate 10, 11 and 12 copies with orange, blue and green, respectively.
Fig. 4.—
(A) Linkage disequilibrium (LD) in and around FLG gene. The x-axis of this plot shows the chromosomal location. The y-axis shows the LD R2 between all reported single nucleotide variants in CHB population and rs77422831, which is the variant with highest iHS score in CHB population. Top panel shows the approximate locations of genes, including FLG and HRNR. The red to blue color gradient indicates decreasing LD with rs77422831.
Analysis of ancient genomes reveal that the Huxian haplogroup is among the genetic variants carried by the first immigrants out-of-Africa. It appears that the Ust’-Ishim genome (Fu et al. 2014), which is from a 45,000-year-old Siberian individual was heterozygously carrying Huxian haplogroup. Maybe more surprisingly, an analysis of ancient European genomes (Haak et al. 2015) revealed that the Huxian haplogroup seems to be absent in Western European Hunter Gatherers, quickly increasing to the modern day frequencies of ∼10–20% by European Early Neolithic (supplementary table S2, Supplementary Material online). In the dataset we analyzed, there were only six western hunter-gatherer genomes for which only two informative single nucleotide variants that tag Huxian haplogroup. As such, we refrain from making conclusions with regard to this increase in the frequency of the Huxian haplogroup in Europe. However, it is clear that the Huxian haplogroup has had a <20% frequency in European populations since the Early Neolithic (last 5,000 years). It is also clear that the Huxian haplogroup evolved in Africa before the out-of-Africa migrations based on the current intermediate frequency of this haplogroup in extant Africans (3–16%), as well as the presence of this haplogroup in multiple ancient Eurasian genomes, including the 45,000-year-old Ust’-Ishim genome. As such, the most likely explanation of the high frequency and extended linkage disequilibrium observed for this haplogroup in Asian populations is Asia-specific adaptive forces acting on standing variation.
We then resolved the copy number variation of the filaggrin repeats within FLG. Previous studies documented that the copy number of subexonic filaggrin repeats can vary from 10 to 12 copies in human populations and dose-dependent correlation between reduced atopic dermatitis risk and the copy number of filaggrin repeats in Irish (Brown et al. 2012), Korean (Li et al. 2016) and African American populations (Quiggle et al. 2015). Since the current pipeline for constructing the 1-kGP dataset was not able to detect subexonic FLG copy number variants (CNVs), we used long-range polymerase chain reaction to genotype these variants in 126 samples from different ancestral backgrounds that are included in 1-kGP dataset (supplementary fig. S5A and table S3, Supplementary Material online). As previously reported (Gan et al. 1990), we found that the copy number of subexonic repeats within FLG vary from 10 to 12 copies. Our results showed remarkable population specificity of subexonic repeat number, with African haplotypes carrying primarily 10 repeats (73%), European haplotypes primarily carrying 11 repeats (49%), and East Asian haplotypes primarily carrying 12 repeats (63%) (fig. 4D and supplementary fig. S5B, Supplementary Material online). In a separate, smaller sample set, we observed that the indigenous Alaskans are more similar in their copy number allele frequencies to Europeans, rather than Asians (supplementary table S3, Supplementary Material online). When we superimposed the copy number onto the tree, we detected at least one instance of recurrence of 10 copy alleles, shuffling the CNVs across different haplotype backgrounds.
The Integration of Different Functional Variants Reveals Multiple Putative Functional Consequences of the Adaptive Sweep of the Huxian Haplogroup in Asia
To further understand the functional consequences of the Huxian haplogroup in Asia, we searched for functional variants that are linked to this haplogroup. Specifically, we first identified all the Phase 3 single nucleotide variants that are in high linkage disequilibrium (R2 > 0.8) with rs77422831, the defining SNP for the Huxian haplogroup in the CHB population. Then, we searched the genome-wide association and expression quantitative trait loci databases for variants. In parallel, we retrieved Combined Annotation Dependent Depletion Scores loss-of-function and nonsynonymous variants. Overall, our results showed that the Huxian haplogroup influences multiple functional sequences with well-documented medical consequences, but with largely unexplored evolutionary and mechanistic implications (supplementary table S4, Supplementary Material online).
Briefly, we found that all 105 Huxian haplotypes in CHB population carry FLG with 12 filaggrin copies, whereas all 101 nonHuxian haplotypes in this population carry FLG with 11 or 10 filaggrin copies. The selective sweep associated with this haplogroup in CHB population explains the observed high frequency of 12 filaggrin-copy alleles of FLG in Asian populations. The expectation, based on the protective effect of high filaggrin copy number alleles against atopic dermatitis in Irish population (Brown et al. 2012), is that the Huxian haplogroup would also be protective against atopic dermatitis in the CHB population. In contrast, we found that all CHB Huxian haplotypes carry the derived single nucleotide variant, rs3126085, which is the top susceptibility variant with a strong effect size for atopic dermatitis susceptibility in Asian populations (Zheng et al. 2011). Indeed, further scrutinization revealed that all Asian Huxian haplotypes carry the FLG2 LoF variant, rs12568784, which has also been associated with increased susceptibility to atopic dermatitis (Margolis, Gupta, Apter, Ganguly, et al. 2014). In addition, we found that the Huxian haplotypes from CHB population carry 21 nonsynonymous FLG SNPs. These multiple and occasionally conflicting functional implications of the Huxian haplogroup to FLG and FLG2 function and its contrasting predicted effect to atopic dermatitis susceptibility lent further support to our hypothesis that the FLG/FLG2 functional variation is evolving neutrally and atopic dermatitis may not alter fitness in human populations to an observable degree. Instead, the significant iHS signature we observed may have been caused by a variant affecting the function of another gene with distinct phenotypic consequences.
One candidate for the target of the selective sweep increasing the frequency of the Huxian haplogroup in Asian populations is the hornerin (HRNR) gene. HRNR is immediately downstream of FLG and is covered by the haplotype block that defines the Huxian haplogroup. As another member of the epidermal differentiation complex, the structure of the gene is similar to FLG and FLG2, harboring large subexonic repeats. However, unlike FLG and FLG2, very few and rare LoF variants affect HRNR (fig. 3D and supplementary table S1, Supplementary Material online). Not much is known about the function of this gene. Nevertheless, recent studies have shown that HRNR is expressed in skin along with other epidermal differentiation complex genes (Wu et al. 2009; Henry et al. 2011), but mostly during wound healing or in psoriatic skin (Takaishi et al. 2005). This gene has also been shown to be actively expressed in tissues other than skin (Fleming et al. 2012).
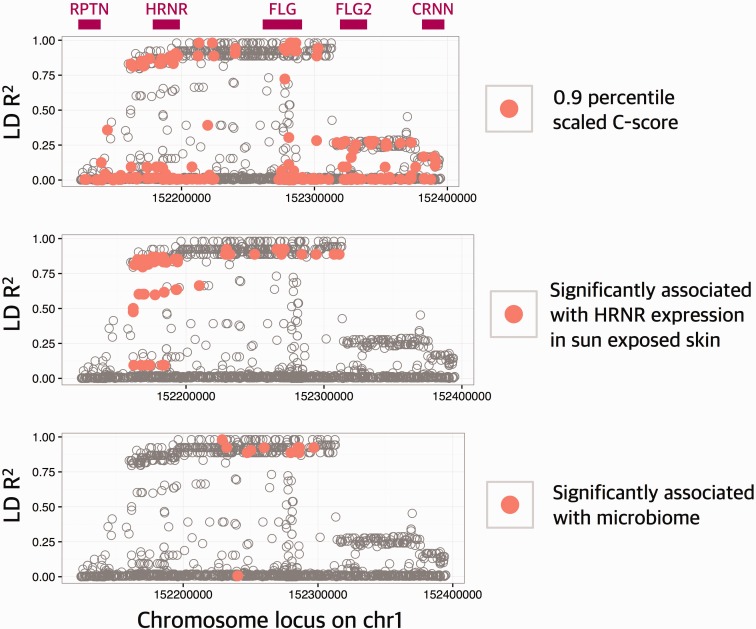
To understand the putative functional impact of Huxian haplogroup, we conducted three analyses (fig. 5 and supplementary table S4, Supplementary Material online). First, we used Combined Annotation Dependent Depletion (CADD) score, which measures putative function of single nucleotide variants in the human genome (Kircher et al. 2014) across the locus to show that multiple putatively functional variants strongly linked with the Huxian haplotype. These include 8 nonsynonymous HRNR single nucleotide variants, separating these haplotypes from nonHuxian haplotypes with unknown functional consequences. Second, we used GTEx database (The GTEx Consortium 2015) to reveal multiple significant expression quantitative variants (P < 10−5), which are strongly linked with Huxian haplogroup in skin. The effect size of the Huxian haplotype is large, but negative, reducing the HRNR expression in >50% in multiple tissues (supplementary fig. S6, Supplementary Material online). We also noticed that Huxian haplotype significantly (P < 10−5) increase the expression of previously reported long noncoding RNA (FLG-AS1). This antagonistic effect is interesting, and potentially imply a RNA level suppression of HRNR expression through a FLG-AS1 intermediary. Third, we conducted an analysis of association between genetic variation in this locus and microbiome composition in multiple tissues using data and methods described in a recent study (Blekhman et al. 2015). We found that single nucleotide variants that are carried by Huxian haplotype are significantly associated with the first principal component of the bacterial abundance diversity in the retroauricular crease (i.e., the skin at the back of the ear) (supplementary table S4, Supplementary Material online). This is especially important in light of recent studies on important role for human genetic variation in shaping the composition of the microbiome (Benson et al. 2010; Goodrich et al. 2014; Blekhman et al. 2015; Davenport et al. 2015).
Fig. 5.—
Putatively functional variants are linked with Huxian haplogroup. The x-axis of this plot shows the chromosomal location. The y-axis shows the LD R2 between all reported single nucleotide variants in CHB population and rs77422831, which is the defining variant for Huxian haplogroup. Top panel shows the approximate locations of genes. In each panel, a different functional aspect of the variation was considered. In the top panel, the variants with CADD scores in the 90th percentile (CADD = 6.714355) were indicated with red points. In the middle panel, the variants that are significantly (P < 10−5) associated with the expression levels of HRNR were indicated with red points. In the bottom panel, the variants that are associated with skin microbiome diversity (P < 0.001) were indicated by red points.
This multilayered functional impact of Huxian haplogroup may imply a complicated evolutionary mechanism involving effect of multiple genes in the epidermal differentiation complex, pathogenic pressures, as well as other physical factors in a given environment, the activity of the immune system of the host, and the microbiome of the skin.
Conclusion
One of the main questions of evolutionary medicine is why some common variants that confer to disease susceptibility remain in the population (Stearns 2012). If we assume that human diseases negatively affect fitness, then we would expect that the disease-susceptibility variants should be eliminated from the population with the effect of purifying (negative) selection. However, there are thousands of relatively common disease-susceptibility variants in modern human populations. Most frequently, the explanation for this observation is that the effect of selection is dwarfed by the effect of genetic drift in human populations (Keinan and Clark 2012). There is, however, a small portion of common genetic variants with relatively large phenotypic effects that still remain in the population, as observed by the FLG LoF variants. FLG is one of the primary loci that has been associated with atopic dermatitis susceptibility in multiple populations with effect sizes for variants linked with FLG reaching I2 = 62% (EArly Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium et al. 2015). Its high levels of expression in the skin tissue and relatively well characterized role in epidermis development are all concordant with the epidemiological implications of the functional variation affecting this gene. As such, from an evolutionary point of view, it is rather unexpected to find a high frequency of LoF mutations affecting this gene, even considering the low effective population sizes of human populations. Based on our results, the most plausible explanation is that despite the negative effects of atopic dermatitis on human well-being, the fitness advantage of not being susceptible to this disease may not be very high. In fact, that extant chimpanzees carry LoF variants of this gene as well. Therefore, the most likely conclusion is that FLG has been accumulating LoF variants since human chimpanzee ancestor, and the current distribution of these variants is primarily shaped by genetic drift.
Even if the effects of negative selection are relaxed for FLG, the functional variation affecting this gene is connected to variation affecting other genes physically and functionally. When we considered physical connections, we discovered a haplogroup that is defined by dozens of putative functional variants affecting the functions of FLG and HRNR. This haplogroup also carries the primary susceptibility variant in Asians, and is the primary contributor to a significant signal of the selective sweep observed in Chinese population. A combination of these physically linked variants may have contributed to atopic dermatitis susceptibility, as well as other allergic reactions known to co-occur with atopic dermatitis, such as rheumatoid arthritis and inflammatory bowel disease (Schmitt et al. 2016). In addition, it is likely that a currently unknown adaptive force acting on one of these variants is the primary driver for the observed selective sweep. The likely target of this adaptive force is the HRNR function. Unlike FLG, HRNR does not harbor common LoF variants. It is plausible, therefore, that one of the nonsynonymous or regulatory variants affecting HRNR are the main target for the selective sweep and that the atopic dermatitis risk variants hitchhiked this sweep, increasing the haplogroup frequency in Asian populations. Such phenomenon may be relatively frequent across the genome, especially in loci with low levels of recombination.
The evolutionary impact of the Huxian haplogroup gets even more complicated when we consider the antagonistic epidemiological effects reported for the variation in the epidermal differentiation complex on chromosome 1, where FLG also resides. Specifically, the common skin diseases, atopic dermatitis and psoriasis have been shown to be affected primarily by epidermal differentiation and immune response. However, recent studies have shown that psoriasis and atopic dermatitis co-occur much less often than expected by chance, and, in fact, there are multiple genetic variants that have opposing effects even though they influence the same biological pathways (Baurecht et al. 2015). Our observation that the Huxian haplogroup is significantly associated with decreased HRNR expression also hints at expression level regulation in the epidermal differentiation complex, with unknown phenotypic effects. May be even more importantly, we showed that Huxian haplogroup is significantly associated with microbial variation. This contributes to the emerging notion that loci with immune or barrier functions interact with the microbiome in various tissues, leading to important biomedically relevant phenotypes, including autoimmune disorders (Ruff and Kriegel 2015; Chu et al. 2016).
Overall, our study leveraged recently available genomic databases to reveal the complex evolutionary history of one of the major disease susceptibility loci. By doing so, our study raises two questions. First, of the many functional variants that the Huxian haplogroup carries, which one confers susceptibility to atopic dermatitis? Second, what is the selective force and its target that drives the selective sweep in Asian populations? For answering both of the questions, it is important to characterize the distinct functions of the structurally similar FLG, FLG2 and HRNR genes. Our locus-specific approach is applicable to many other disease-associated loci with complex evolutionary histories and will expand our understanding of the evolutionary reasons behind disease susceptibility.
Methods
Samples and Detection of Nonsense (Stop-Gain or Loss-of-Function) Single Nucleotide Variants
We used 1000 Genomes Phase 3 dataset (1-kGP dataset, http://www.1000genomes.org/announcements/initial-phase-3-variant-list-and-phased-genotypes-2014-06-24), which involve resequencing data from 2,504 samples across 26 global populations (1000 Genomes Project Consortium et al. 2015) for all our analyses. We used nonsense single nucleotide variants from dbSNP track (Human built 142) from Table Browser of the UCSC Genome Browser (Karolchik et al. 2004). Using the obtained list of nonsense SNPs we employed python code to discern about 51 nonsense SNPs in FLG from the 1-kGP phase 3 dataset. We manually verified for the stop-gain function of each FLG LoF variant. Following a similar method, we also extracted nonsense variants in the other SFTP genes including trichohyalin-like 1 (TCHHL1), trichohyalin (TCHH), repetin (RPTN), hornerin (HRNR), filaggrin-2 (FLG2) and cornulin (CRNN). For confirmation, we downloaded all the stop-gain variants from the ExAC database and also manually searched for each 1000 Genomes LoF variant in this dataset to curate supplementary table S1, Supplementary Material online.
Detection of CNVs
We used long-range PCR method, slightly modified from Brown et al. (2012), to amplify the 3′ portion of FLG exon 3, spanning repeats 7–10 including 3′ partial repeat region. A total of 126 individuals representing different global populations were included for this analysis (supplementary table S3, Supplementary Material online). Briefly, the long-range PCR reaction was performed using Expand High Fidelity System kit (Roche Applied Science, Mannheim, Germany). The PCR reactions (20 μl) was prepared from 2 μl of the Buffer2 (with MgCl2), 2 μl of dNTPs (2.5 mM each), 10 μM each of the forward (5′-CCCAGGACAAGCAGGAACT-3′) and reverse (5′-CTGCACTACCATAGCTGCC-3′) primers, DMSO (4% v/v), 0.35 μl of Expand High Fidelity enzyme, ∼100–200 ng of the genomic DNA and molecular grade water. Thermal cycle conditions were as follows: 95 °C initial denaturation for 5 min, followed by 35 cycles of denaturation (94 °C for 30 s), annealing (64.3 °C for 30 s) and elongation (72 °C for 5 min 30 s), followed by a final elongation at 72 °C for 7 min. The product size (4,277 bp—10 copies; 5,249 bp—11 copies; 6,224 bp—12 copies) were identified using gel electrophoresis by running the PCR products along with 1-kb ladder on a 0.8% w/v agarose gel (supplementary fig. S5A, Supplementary Material online), at 120 V for 1–2 h.
Population Genetic Analysis
We constructed the maximum likelihood tree for the 10,000-bp upstream region of the FLG gene based on the 1-kGP phase 3 dataset using RAxML version 8.0.0 (Stamatakis 2014). We then manually detected major branches of the tree and calculated the number of haplotypes in each branch in continental populations. For understanding the co-occurrence of functional single variants on individual haplotypes, we curated 55 SNPs that included 51 FLG LoF variants, the FLG2 LoF variant (rs12568784), the top iHS variant in CHB and CEU populations (rs77422831, iHS in CHB: 2.75625, iHS in CEU: 3.5683), and the significant genome wide association study variant (rs12568784). Our results found 96 distinct haplotypes, most of which are rare. We plotted the common variants using R rworldmap package. We obtained integrated haplotype homozygosity scores (iHS), FST and nucleotide diversity (pi) values for CEU, CHB and YRI from 1000 Genomes Selection Browser 1.0 database (http://hsb.upf.edu, last accessed December 2015). For finding the minor allele frequency for Bronze Age Europeans, we used the dataset from Haak et al. (2015). The genotypes were extracted using EIGENSOFT (v6.0.1) (Patterson et al. 2006; Price et al. 2006). The Python and R codes used for calculating the frequency and generating plots are available online (http://gokcumenlab.org/data-and-codes/).
Functional Analyses
We used the CADD tool (Kircher et al. 2014) to get the CADD-based C-scores that gives the deleteriousness of the single nucleotide variants of the 1000 Genome Phase 3 dataset (http://cadd.gs.washington.edu/download). Compared with the raw C-scores, we used scaled C-scores, which is “PHASED-scaled” based on the rank of all ∼8.6 billion single nucleotide variants of the GRCh37/hg19 reference genome. We calculated linkage disequilibrium between chosen SNPs and all structural variants reported in 1000 Genomes Phase 3 dataset. The relevant python codes used for calculating linkage disequilibrium are available online (http://gokcumenlab.org/data-and-codes/).
For expression quantitative trait loci analysis, we searched rs12746538, one SNP linked with Huxian haplogroup, in the GTEx database (http://www.gtexportal.org/home/) (The GTEx Consortium 2015). Our results showed that this variant is significantly associated with the expression levels of FLG and HRNR genes in multiple tissues.
For the microbiome data, we used the approach and the data described in Blekhman et al. (2015). Briefly, the microbiome 16s data from 15 different body sites were used as quantitative traits to construct principal components. Then, individual principal components were correlated against the host genetic variation.
Statistical Analysis and Graphs
All statistical analysis including linear regression (R2), Wilcoxon rank sum test (P value), Fisher’s exact test (P value) and graphs were performed using basic statistical test and ggplot2 packages available through R version 3.2.1 (https://www.r-project.org). All codes used in this study have been provided in our website: http://gokcumenlab.org/data-and-codes/.
Supplementary Material
Supplementary figures S1–S6 and table S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Authors Contributions
O.G., M.E., N.J., M.G.H., and R.B. wrote the main article and designed the experiments. M.E., D.X., and O.G. prepared figures. M.E., C.F., and M.R. conducted PCR for copy number variation genotyping. M.E. and D.X. conducted most bioinformatic analyses. R.B. conducted microbiome analyses. All authors reviewed the article.
Supplementary Material
Acknowledgments
We thank Derek Taylor for his comments on earlier versions of this article. This study is supported by OG’s start-up funds from University at Buffalo Research Foundation. NSF OPP-0732857 supported genotyping of the Alaskan North Slope samples.
Literature Cited
- 1000 Genomes Project Consortium, et al. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova-Fischer I, et al. 2011. Distinct barrier integrity phenotypes in filaggrin-related atopic eczema following sequential tape stripping and lipid profiling. Exp Dermatol. 20:351–356. [DOI] [PubMed] [Google Scholar]
- Barresi C, et al. 2011. Increased sensitivity of histidinemic mice to UVB radiation suggests a crucial role of endogenous urocanic acid in photoprotection. J Invest Dermatol. 131:188–194. [DOI] [PubMed] [Google Scholar]
- Baurecht H, et al. 2015. Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am J Hum Genet. 96:104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 107:18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, et al. 2015. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16:191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, et al. 2012. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol. 132:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, McLean WHI. 2012. One remarkable molecule: filaggrin. J Invest Dermatol. 132:751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 6:328–340. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. 2011. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br J Dermatol. 165:106–114. [DOI] [PubMed] [Google Scholar]
- Chu H, et al. 2016. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352(6289):1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. 2005. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15:1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale BA, Holbrook KA, Steinert PM. 1978. Assembly of stratum corneum basic protein and keratin filaments in macrofibrils. Nature 276:729–731. [DOI] [PubMed] [Google Scholar]
- Davenport ER, et al. 2015. Genome-wide association studies of the human gut microbiota. PLoS One 10:e0140301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cid R, et al. 2009. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 41:211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EArly Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium, Australian Asthma Genetics Consortium (AAGC), Australian Asthma Genetics Consortium AAGC. 2015. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 47:1449–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JM, Ginsburg E, Oliver SD, Goldsmith P, Vonderhaar BK. 2012. Hornerin, an S100 family protein, is functional in breast cells and aberrantly expressed in breast cancer. BMC Cancer 12:266.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan SQ, McBride OW, Idler WW, Markova N, Steinert PM. 1990. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry 29:9432–9440. [DOI] [PubMed] [Google Scholar]
- Gao P-S, et al. 2009. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 124:507–513, 513.e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokcumen O, et al. 2013. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci U S A. 110:15764–15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, et al. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, et al. 2011. Filaggrin genotype in ichthyosis vulgaris predicts abnormalities in epidermal structure and function. Am J Pathol. 178:2252–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J, et al. 2011. Hornerin is a component of the epidermal cornified cell envelopes. FASEB J. 25:1567–1576. [DOI] [PubMed] [Google Scholar]
- Holick MF, Chen TC, Lu Z, Sauter E. 2007. Vitamin D and skin physiology: a D-lightful story. J Bone Miner Res. 22(Suppl 2):V28–V33. [DOI] [PubMed] [Google Scholar]
- Hsu C-K, et al. 2009. Analysis of Taiwanese ichthyosis vulgaris families further demonstrates differences in FLG mutations between European and Asian populations. Br J Dermatol. 161:448–451. [DOI] [PubMed] [Google Scholar]
- Jablonski NG. 2012. Human skin pigmentation as an example of adaptive evolution. Proc Am Philos Soc. 156:45–57. [PubMed] [Google Scholar]
- Jablonski NG, Chaplin G. 2000. The evolution of human skin coloration. J Hum Evol. 39:57–106. [DOI] [PubMed] [Google Scholar]
- Karolchik D, et al. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Clark AG. 2012. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336:740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, et al. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraning KK. 1991. Temperature regulation and the skin In: Goldsmith LA, editor. Physiology, biochemistry, and molecular biology of the skin. New York: Oxford University Press; p. 1085–1095. [Google Scholar]
- Kypriotou M, Huber M, Hohl D. 2012. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the “fused genes” family. Exp Dermatol. 21:643–649. [DOI] [PubMed] [Google Scholar]
- Lachance J, Tishkoff SA. 2013. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. Bioessays 35:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, et al. 2016. Copy-number variation of the filaggrin gene in Korean patients with atopic dermatitis: what really matters, “number” or “variation”? Br J Dermatol. 174:1098–1100. [DOI] [PubMed] [Google Scholar]
- Lin Y-L, Pavlidis P, Karakoc E, Ajay J, Gokcumen O. 2015. The evolution and functional impact of human deletion variants shared with archaic hominin genomes. Mol Biol Evol. 32(4):1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, et al. 2007. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat Genet. 39:1261–1265. [DOI] [PubMed] [Google Scholar]
- Margolis DJ, Gupta J, Apter AJ, Ganguly T, et al. 2014. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J Allergy Clin Immunol. 133:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Gupta J, Apter AJ, Hoffstad O, et al. 2014. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J Invest Dermatol. 134:2272–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner M, et al. 2010. Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. J Invest Dermatol. 130:2286–2294. [DOI] [PubMed] [Google Scholar]
- Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. 1996. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol. 106:989–992. [DOI] [PubMed] [Google Scholar]
- Palmer CNA, et al. 2006. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 38:441–446. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presland RB, Haydock PV, Fleckman P, Nirunsuksiri W, Dale BA. 1992. Characterization of the human epidermal profilaggrin gene. Genomic organization and identification of an S-100-like calcium binding domain at the amino terminus. J Biol Chem. 267:23772–23781. [PubMed] [Google Scholar]
- Price AL, et al. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38:904–909. [DOI] [PubMed] [Google Scholar]
- Proksch E, Brandner JM, Jensen J-M. 2008. The skin: an indispensable barrier. Exp Dermatol. 17:1063–1072. [DOI] [PubMed] [Google Scholar]
- Pybus M, et al. 2014. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42:D903–D909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiggle AM, et al. 2015. Low filaggrin monomer repeats in African American pediatric patients with moderate to severe atopic dermatitis. JAMA Dermatol. 151:557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings AV, Harding CR. 2004. Moisturization and skin barrier function. Dermatol Ther. 17(Suppl 1):43–48. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, et al. 2010. Genome-wide association studies in diverse populations. Nat Rev Genet. 11:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquier S, Blancher A, Giorgi D. 2000. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc Natl Acad Sci U S A. 97:2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff WE, Kriegel MA. 2015. Autoimmune host–microbiota interactions at barrier sites and beyond. Trends Mol Med. 21:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands A, et al. 2007. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 39:650–654. [DOI] [PubMed] [Google Scholar]
- Schmitt J, et al. 2016. Atopic dermatitis is associated with an increased risk for rheumatoid arthritis and inflammatory bowel disease, and a decreased risk for type 1 diabetes. J Allergy Clin Immunol. 137:130–136. [DOI] [PubMed] [Google Scholar]
- Scott IR, Harding CR, Barrett JG. 1982. Histidine-rich protein of the keratohyalin granules: source of the free amino acids, urocanic acid and pyrrolidone carboxylic acid in the stratum corneum. Biochim. Biophys. Acta. 719:110–117. [DOI] [PubMed] [Google Scholar]
- Simon M, et al. 1996. Evidence that filaggrin is a component of cornified cell envelopes in human plantar epidermis. Biochem J. 317 (Pt 1): 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FJD, et al. 2006. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 38:337–342. [DOI] [PubMed] [Google Scholar]
- Somel M, et al. 2013. A scan for human-specific relaxation of negative selection reveals unexpected polymorphism in proteasome genes. Mol Biol Evol. 30:1808–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC. 2012. Evolutionary medicine: its scope, interest and potential. Proc Biol Sci. 279:4305–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Marekov LN. 1995. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 270:17702–17711. [DOI] [PubMed] [Google Scholar]
- Stokowski RP, et al. 2007. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 81: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, et al. 2014. Evolutionary origin and diversification of epidermal barrier proteins in amniotes. Mol Biol Evol. 31:3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi M, Makino T, Morohashi M, Huh N-H. 2005. Identification of human hornerin and its expression in regenerating and psoriatic skin. J Biol Chem. 280:4696–4703. [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen JP, Bikle DD, Elias PM. 2014. Evidence that loss-of-function filaggrin gene mutations evolved in Northern Europeans to favor intracutaneous vitamin D3 production. Evol Biol 41:388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge MCG, Bradley M. 2014. Prevalence of filaggrin gene mutations: an evolutionary perspective In: Thyssen JP, Maibach HI Filaggrin. Berlin Heidelberg: Springer; p. 119–128. [Google Scholar]
- Winge MCG, et al. 2011. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. Br J Dermatol. 165:1074–1080. [DOI] [PubMed] [Google Scholar]
- Wu Z, et al. 2009. Highly complex peptide aggregates of the S100 fused-type protein hornerin are present in human skin. J Invest Dermatol. 129:1446–1458. [DOI] [PubMed] [Google Scholar]
- Zheng XD, et al. 2011. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nature Genetics 43:690–694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.